Report card for Correlation and Hierarchical Mappings on the Human Basal Ganglia ( Johansen, Fu et al 2025)
The consensus basal ganglia taxonomy of Johansen, Fu et al 2025 integrates HMBA single-nucleus RNA sequencing (snRNA-seq) data from human, macaque, marmoset, and previously published mouse basal ganglia, with the goal of generating a consensus cell type taxonomy that can be widely adopted by the scientific community. By focusing on conserved marker genes and shared molecular profiles to supplement established names from the broader community, we have developed a standardized naming system that captures the evolutionary relationships and functional distinctions among basal ganglia cell types. The HMBA consensus basal ganglia taxonomy is designed to streamline communication, foster collaboration, and facilitate the development of novel research tools targeting specific cell types across multiple species.
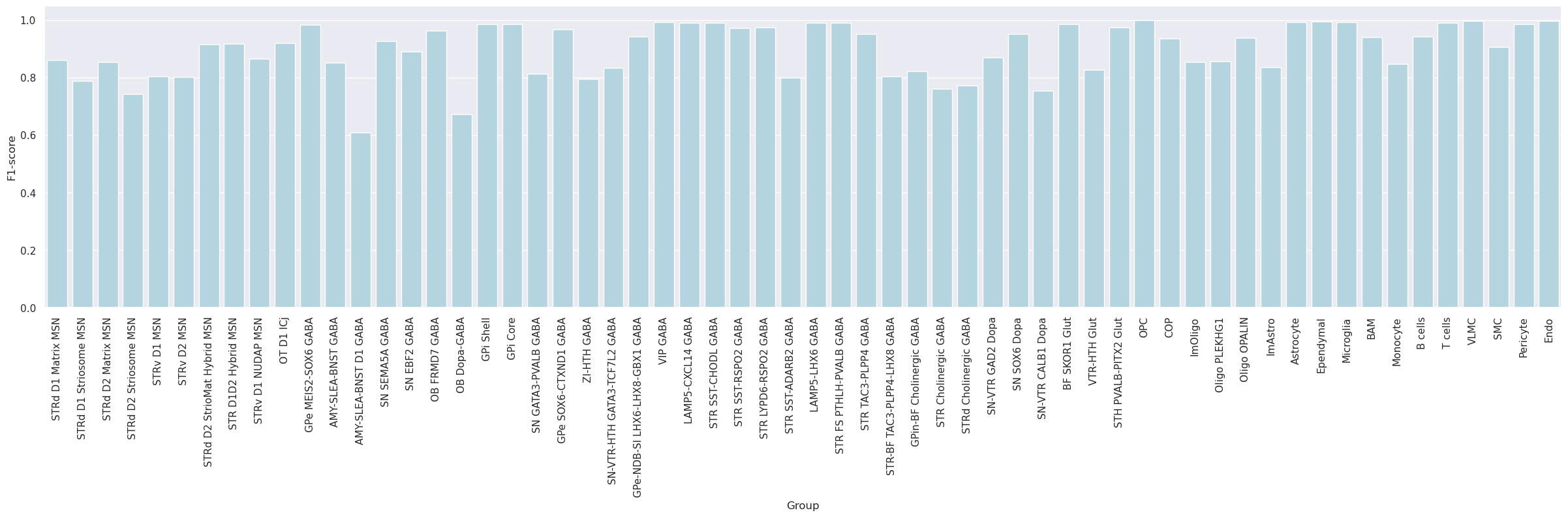
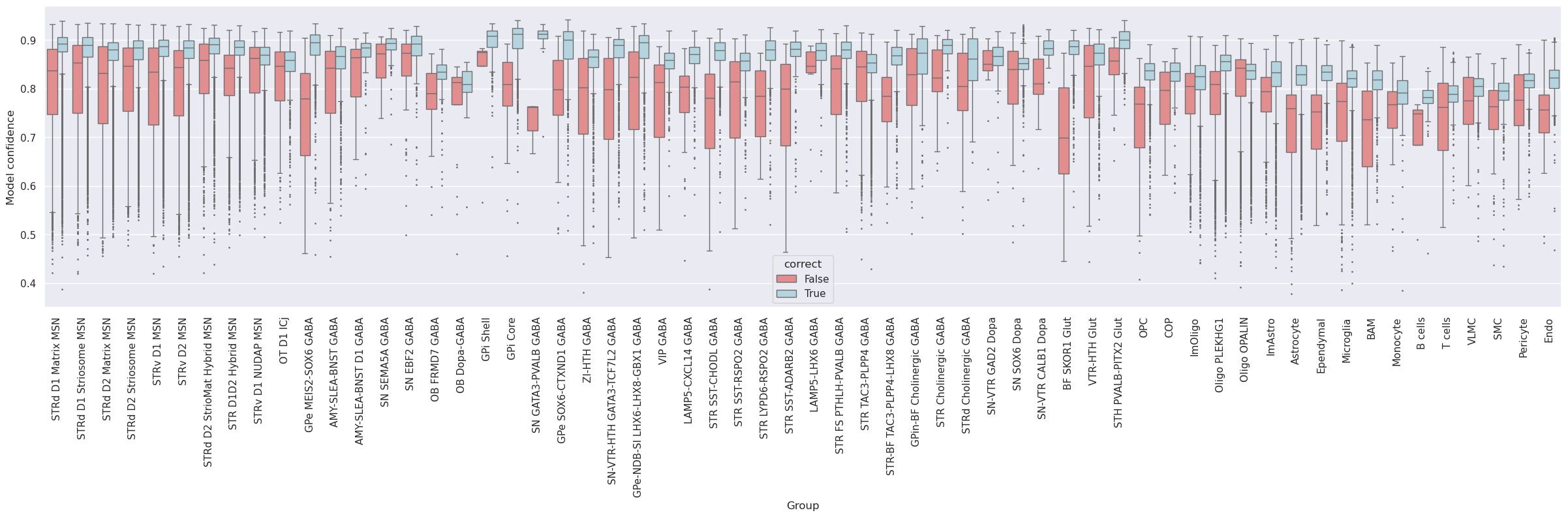
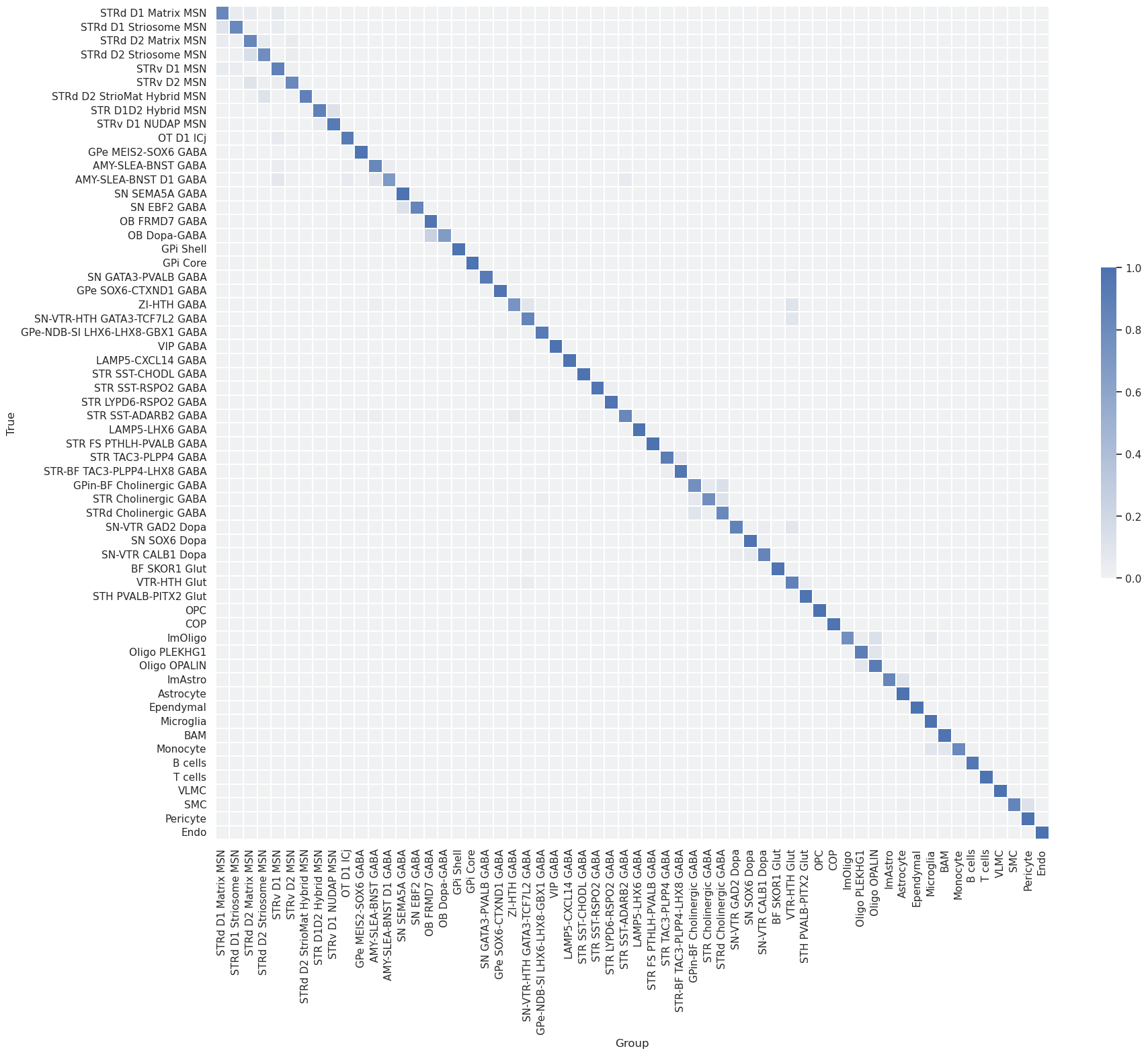
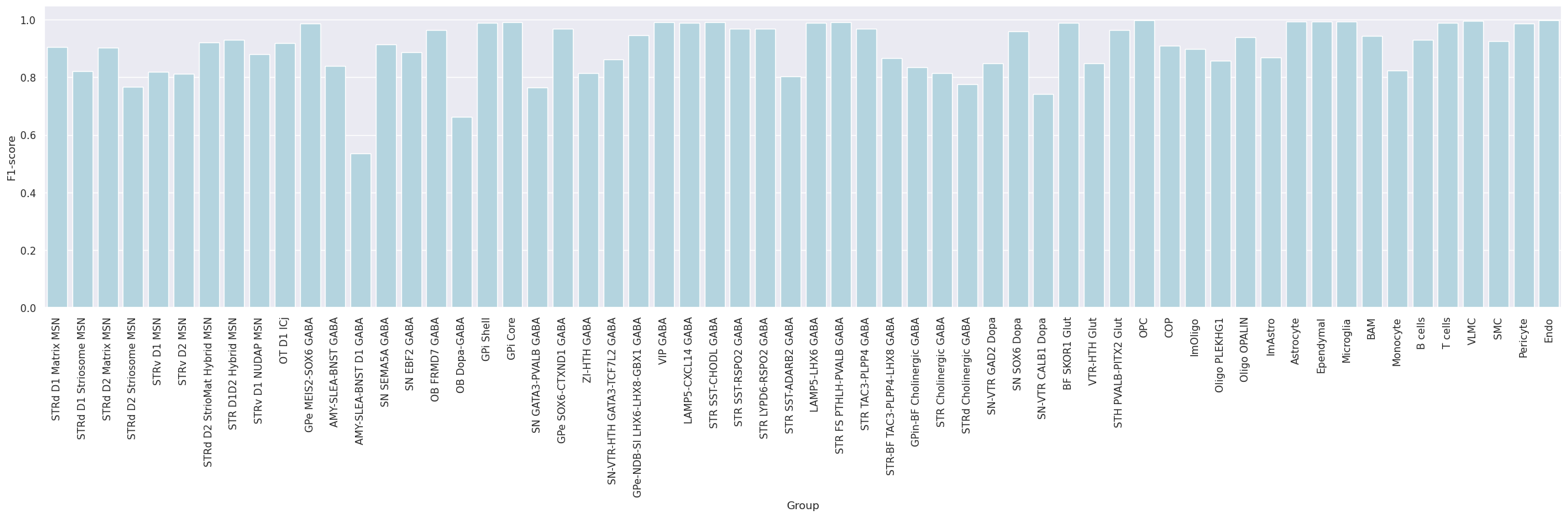
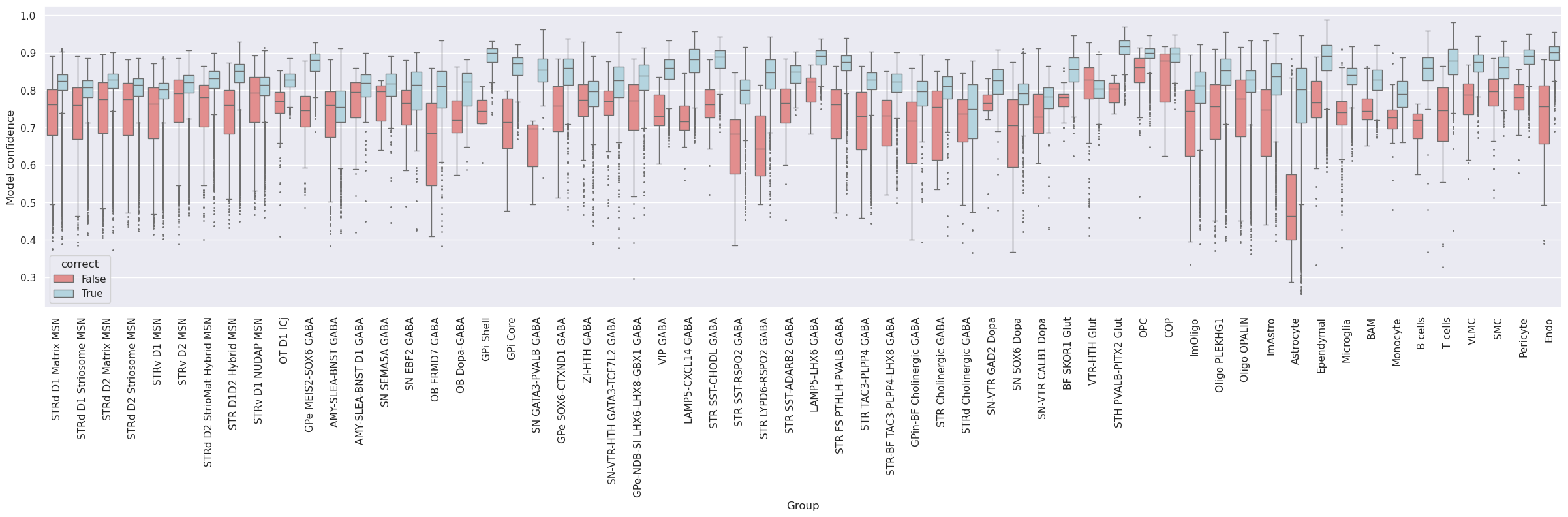
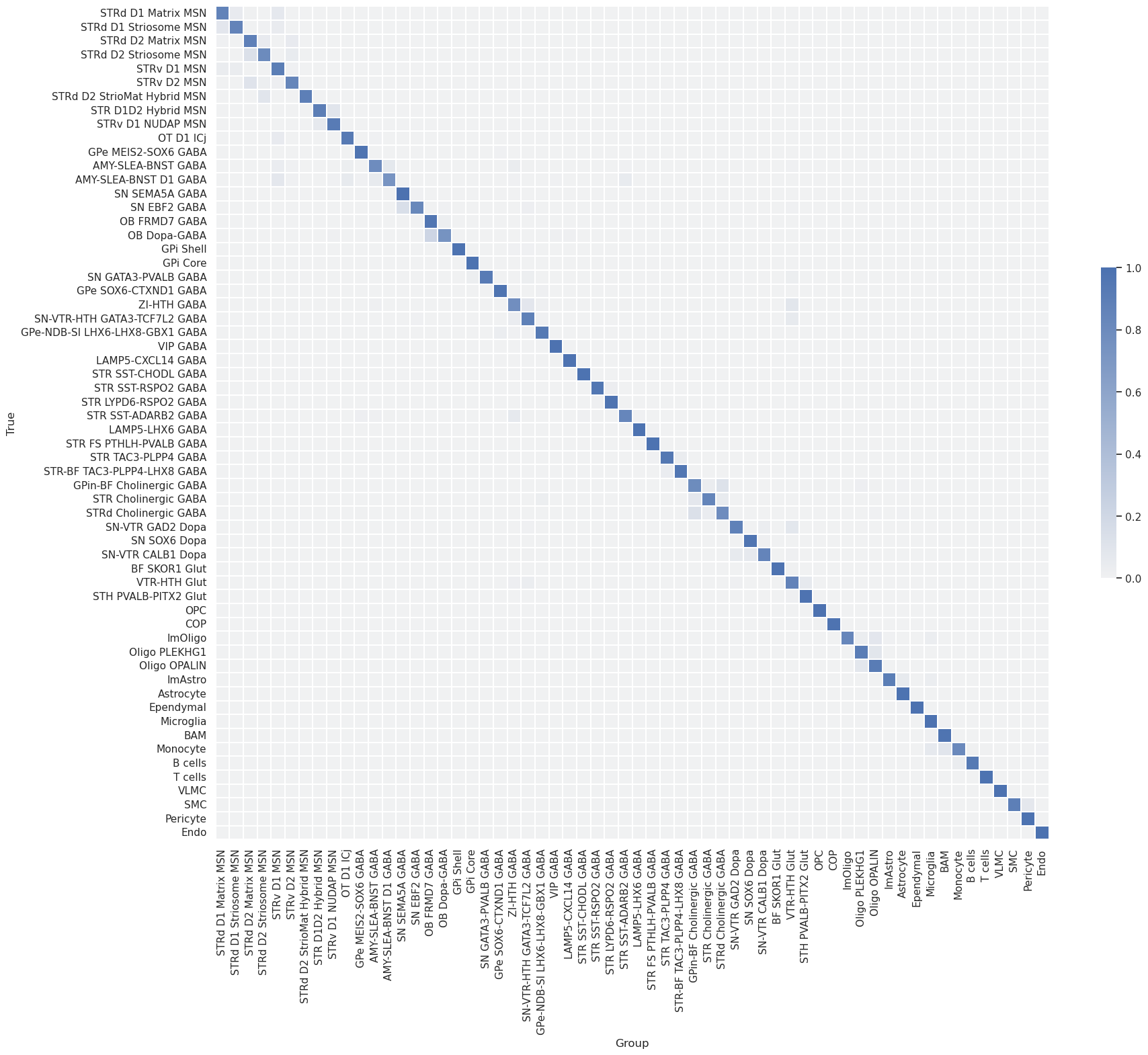
Subsequently, the dataset was mapped to itself, termed self-projection, for evaluating the ideal performance of correlation and hierarchical mapping algorithms.
The analysis evaluates the predictions of correlation and hierarchical mappings in determining cluster labels in a self-projection evaluation.
Annotation
Correlation F1-score
Hierarchical F1-score
Neighborhood
0.985
0.988
Class
0.983
0.987
Subclass
0.957
0.961
Group
0.898
0.903
Cluster
0.630
0.592