The Allen Institute for Brain Science uses the Patch-seq technique to collect high-quality electrophysiological, morphological and transcriptomic data from single neurons. The resources below enable exploration of these multimodal characterization datasets.
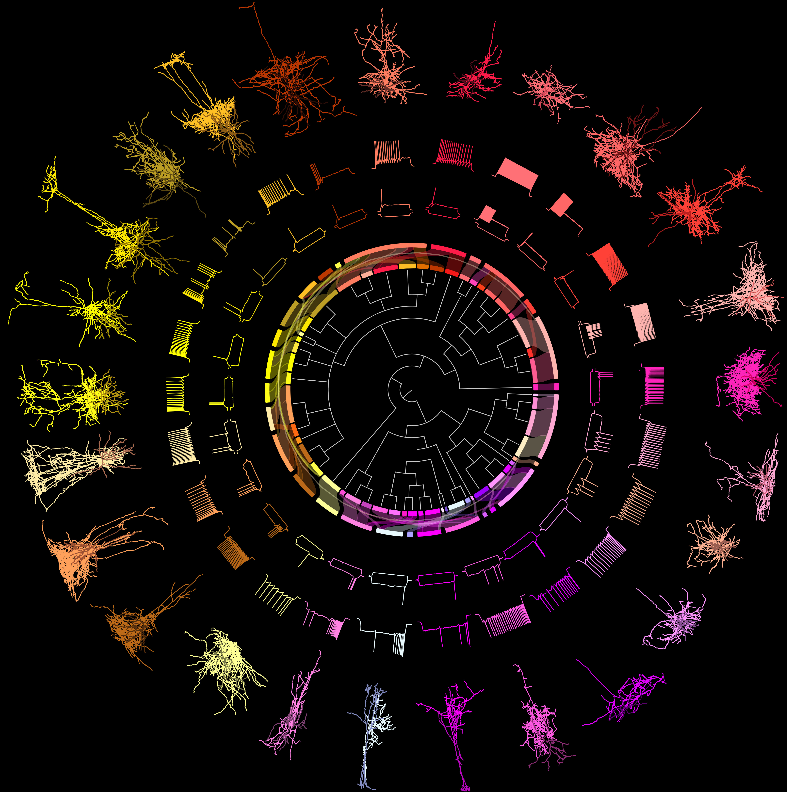

This dataset includes 4,435 mouse cells from the visual cortex. Available data describe the cells' transcriptomes, intrinsic physiological properties, and — for a subset — morphologies. Based on their transcriptomes, these cells were assigned to one of the 60 transcriptomically defined cell types described in Tasic et al. 2018. Nearly all of the cells in this dataset are inhibitory, but includes some excitatory cells. Additionally, cells with all three modalities were assigned to one of 28 interneuron MET-types that have congruent morphological, electrophysiological, and transcriptomic properties.
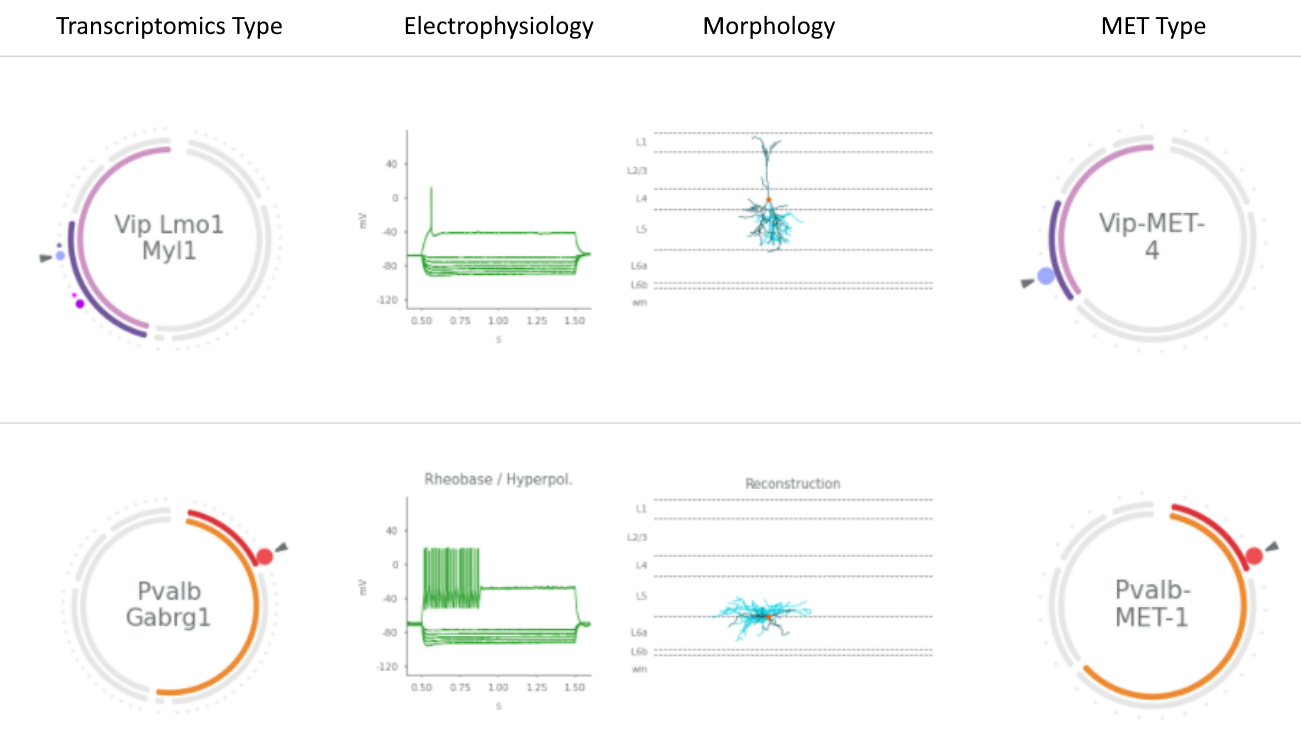
These data facilitate characterization of the morphological and/or intrinsic electrophysiological properties of neurons belonging to a given transcriptomically defined type of GABAergic interneuron. These data also enable examination of the degree to which distinct transcriptomically-defined types of GABAergic interneurons exhibit morphological and/or intrinsic electrophysiological differences. Lastly, the glutamatergic cells from layer 2/3 facilitate cross-species comparison with the human dataset here.
Researchers can use the metadata file to find cells of a particular cell type of interest and then use the manifest file to download raw data files for those cells. Instructions and links are below. This video tutorial provides an overview of the data and walks through an example Jupyter notebook.
Neuroscience Multi-omic Data Archive (NeMO) hosts the transcriptomics data files. This online directory in the NeMO Archive hosts the FASTQ files for this dataset. There are 460 GB of total files.
This online directory in the NeMO Archive hosts the BAM files for this dataset. Note that the checksum values provided in the file manifest are for the files after they have been extracted from the .tar bundle.
This online directory in the NeMO Archive hosts four gene expression matrices: exons, introns, total counts, and normalized counts. Note that the checksum values provided in the file manifest are for the files after they have been extracted from the .tar bundle.
This online directory in Distributed Archives for Neurophysiology Data Integration (DANDI) hosts electrophysiology files. The Download Instructions above include guidance on downloading these data through the DANDI command line.
This online directory in The Brain Image Library (BIL) hosts neuron reconstruction files. BIL supports file downloads using several methods, which are detailed in their instructions here.
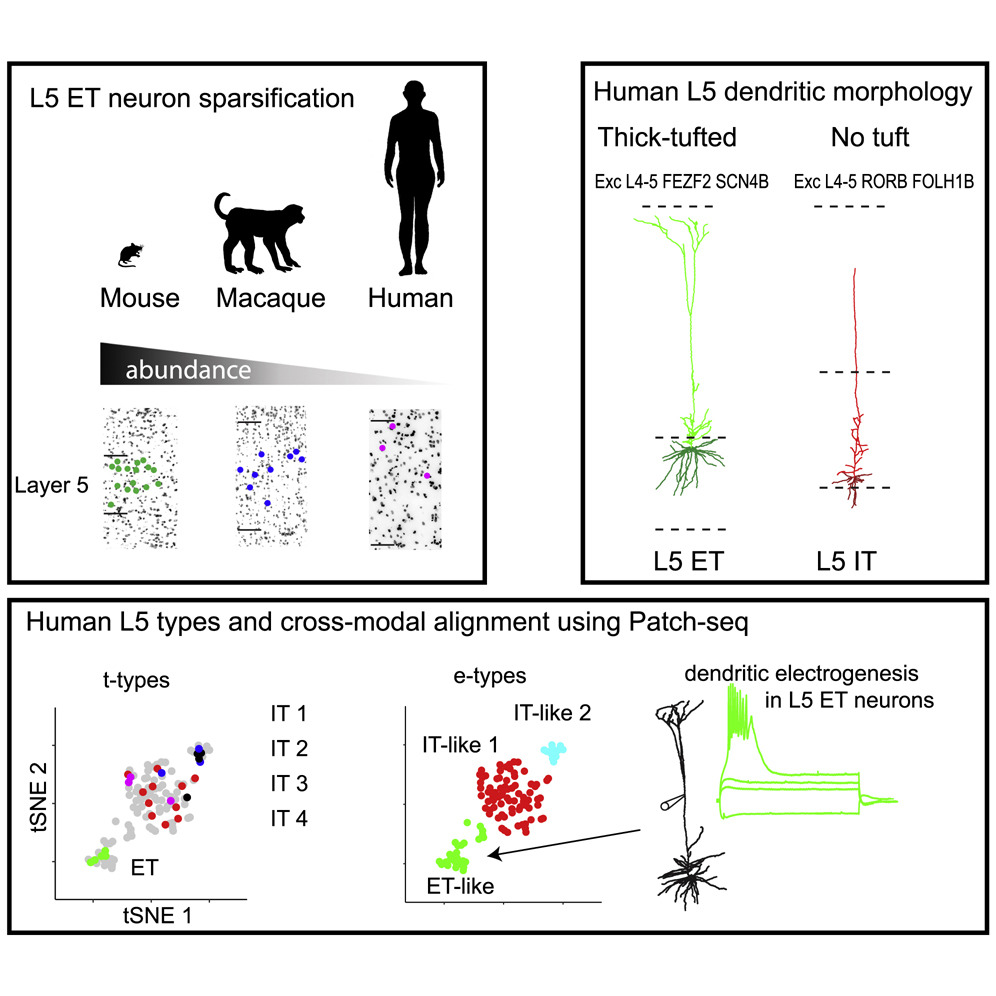

This dataset includes intrinsic membrane properties of 113 human Layer 5 pyramidal neurons acquired via patch-clamp recordings in brain slices from the temporal cortex region. We also collected the transcriptome (n=25) and/or dendritic morphologies (n=15) from a subset of these cells. Together this dataset reveals two broad classes of neurons with transcriptomic and morpho-electric properties resembling extra-telencephalic and intra-telencephalic projecting neurons.
Viral labeling of GABAergic neurons in human brain slices with Patch-seq yields a functional annotation of human interneuron subclasses and types.
Linking cellular transcriptomic identity to intrinsic morphoelectric features, we describe innovations in human neocortical layer 1 interneurons.
We characterized the morphological and physiological properties of five transcriptomically-defined human glutamatergic supragranular neuron types. Three of these types have properties that are specialized as compared to the more homogeneous properties of transcriptomically defined homologous mouse neuron types. The two remaining supragranular neuron types, located exclusively in deep layer 3, do not have clear mouse homologues in supragranular cortex but are transcriptionally most similar to deep layer mouse intratelencephalic-projecting neuron types.
We have optimized the Patch-seq technique to efficiently collect high-quality electrophysiological, morphological and transcriptomic data. Here we describe this optimized technique as well as the publicly available tools that can be used to generate comparable data.