MapMyCells allows users to assign cell type names from Allen Institute-hosted taxonomies to their own single cell and spatial transcriptomics data. It transforms cell types from a concept in publications to a tool for public research.
Such data integration allows users to leverage additional community knowledge and data visualizations in their own research.
MapMyCells transforms cell types from a concept in publications to a tool for public research. Scientists worldwide can discover what cell types their transcriptomics and spatial data corresponds with by comparing their data to massive, high-quality reference datasets.
One key advantage to MapMyCells is scale: using our cloud-based Brain Knowledge Platform and reference datasets with millions of cells, researchers can provide up to 327 million cell-gene pairs from their own data, which is a huge leap forward for working with whole-brain datasets.
MapMyCells will enable the neuroscience community to
Allen Institute does not use, retain, or aggregate any data uploaded to MapMyCells for its own internal purposes, nor will we publish your data publicly. Allen Institute database administrators can access any uploaded dataset for debugging and other error remediation purposes. All files will be deleted one week after upload. Please do not submit any sensitive data, personally identifiable data, or protected health data that could put an individuals' privacy at risk into MapMyCells. See the Allen Institute Privacy Policy for more information on our privacy practices.

A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain.
It contains 5,322 clusters that are organized in a hierarchical manner with nested groupings of 34 classes, 338 subclasses, 1,201 supertypes and 5,322 types/clusters.
A high-resolution transcriptomic atlas of cell types from middle temporal gyrus from the SEA-AD aged human cohort that spans the spectrum of Alzheimer’s disease.
It is hierarchically organized into nested levels of classification: 3 classes, 24 subclasses, and 139 supertypes.

Transcriptomic diversity of cell types in adult human brain.
It is clustered into hierarchical groups of 31 superclusters, 461 clusters, and 3313 subclusters.

A multi-species, multi-omic cell type atlas of the primate basal ganglia that integrates over two million single nuclei from eight major basal ganglia structures profiled using single-nucleus RNA-seq, ATAC-seq, spatial transcriptomics, morphology, and electrophysiology.
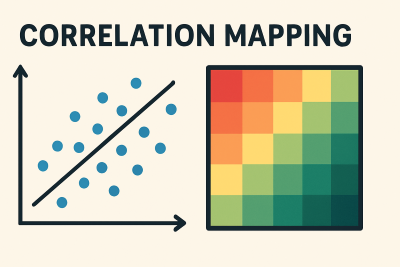


MapMyCells
MapMyCells produces the following output files. A “standard” CSV output file and an “extended” JSON output file. These files are archived into a single .zip file for download.

We recommend first time users start with the step-by-step guide, which describes file formats, use cases, and additional options.
MapMyCells includes a drag-and-drop interface for users to assign cell type names from the above taxonomies to their own data. Files up to 2GB or about 150,000 cells can be processed through the browser.

This code provides a python package for mapping single sell RNA sequencing data onto a cell type taxonomy such as that provided by the Allen Institute for Brain Science.
