Human Mammalian Brain Atlas (HMBA)
Human and Mammalian Brain Atlas (HMBA) is a major component of the BRAIN Initiative Cell Atlas Network (BICAN) that proposes to establish a highly granular cross-mammalian cell atlas, linking brain structure, function, and cellular architecture. The open release of this genome-style achievement, including data, knowledge, and tools, intends to unify how the field identifies and understands brain cell types, accelerating global collaboration and breakthroughs.
First release: Basal Ganglia
The first atlas release from the HMBA consortium delivers a cross-species cell type taxonomy, a massive annotated single-cell dataset, and tools for accessing, exploring, and utilizing the taxonomy and data.


Scientific methods and findings are detailed in a package of pre-prints. Links will be added as they become available in November & December 2025.
- Cross-species consensus atlas of the basal ganglia
- A cross-species spatial transcriptomic atlas of the human and non-human primate basal ganglia
- Spatial patterning of transcriptional and regulatory programs in the primate subcortex
- Morphoelectric diversity and specialization of neuronal cell types in the primate striatum
- Circuit specific specialization of human basal ganglia astrocytes
- Optimized methods for human postmortem brain preparation and 3D CCF mapping
- Towards human and non-human primate common coordinate frameworks using unified structural ontologies
- A consensus spinal cord cell type atlas across mouse, macaque, and human
- Human Neocortical Glutamatergic Neurons Revealed Through Multimodal Profiling
- Aging selectively affects morpho-electric features of intratelencephalic-projecting pyramidal neurons in human brain
- Gene expression and morphological features drive high prevalence of subthreshold membrane potential resonance in human cortical interneurons
- Fast Optimization of Robust Transcriptomics Embeddings using Probabilistic Inference Autoencoder Networks for multi-Omics
HMBA's cross-species cell types taxonomy was also used by other BICAN projects in additional pre-prints
- Molecular zonation of the human striatum across cell types and people
- Inter-individual variation of the adult human striatum
- A Multimodal Single Cell Epigenomic and 3D Genome Atlas of the Human Basal Ganglia
- Single-Cell Analysis of Chromatin State and Transcriptome in Human Basal Ganglia
- Multiscale Spatial Transcriptomic Atlas of Human Basal Ganglia Cell-Type and Cellular Community Organization
- Disruption of Cell-Type-Specific Molecular Programs of Medium Spiny Neurons in Autism
Have a specific question or need more help? Check out our Community Forum
Become part of a supportive community created by scientists for scientists. Find answer to your own questions and help your colleagues use brain-map resources to their full extend with Community Forum's diverse content.
The foundational cell type reference for research communities
The Human and Mammalian Brain Atlas (HMBA) intends to unify how the field identifies and understands brain cell types. Like the human genome project, at the heart of this goal is an open, high-quality, and high-resolution reference for the entire field to use. In the case of HMBA, that reference comprises a cross-mammalian whole-brain cell type taxonomy and a harmonized cross-species anatomical atlas, with tight data driven linkages between the cell types and anatomy. Around the beating heart of the reference, HMBA will provide tools for the community to access underlying data, visualize the taxonomy and atlases and co-visualize their own data, search the references, and map their data to the references. Just how the tool ecosystem for the human genome empowers researches to use the reference genome in their research, the tools from HMBA will empower the neuroscience community to label, analyze, visualize, and interpret their research faster and with greater confidence. When the whole neuroscience field can speak the same language about cell types and anatomy, scientists can collaborate faster and more effectively, allowing more time to design incisive experiments, interpret complex disease mechanisms, and develop better treatments. What the human genome project did for genomics, this atlas aims to do for cell types.
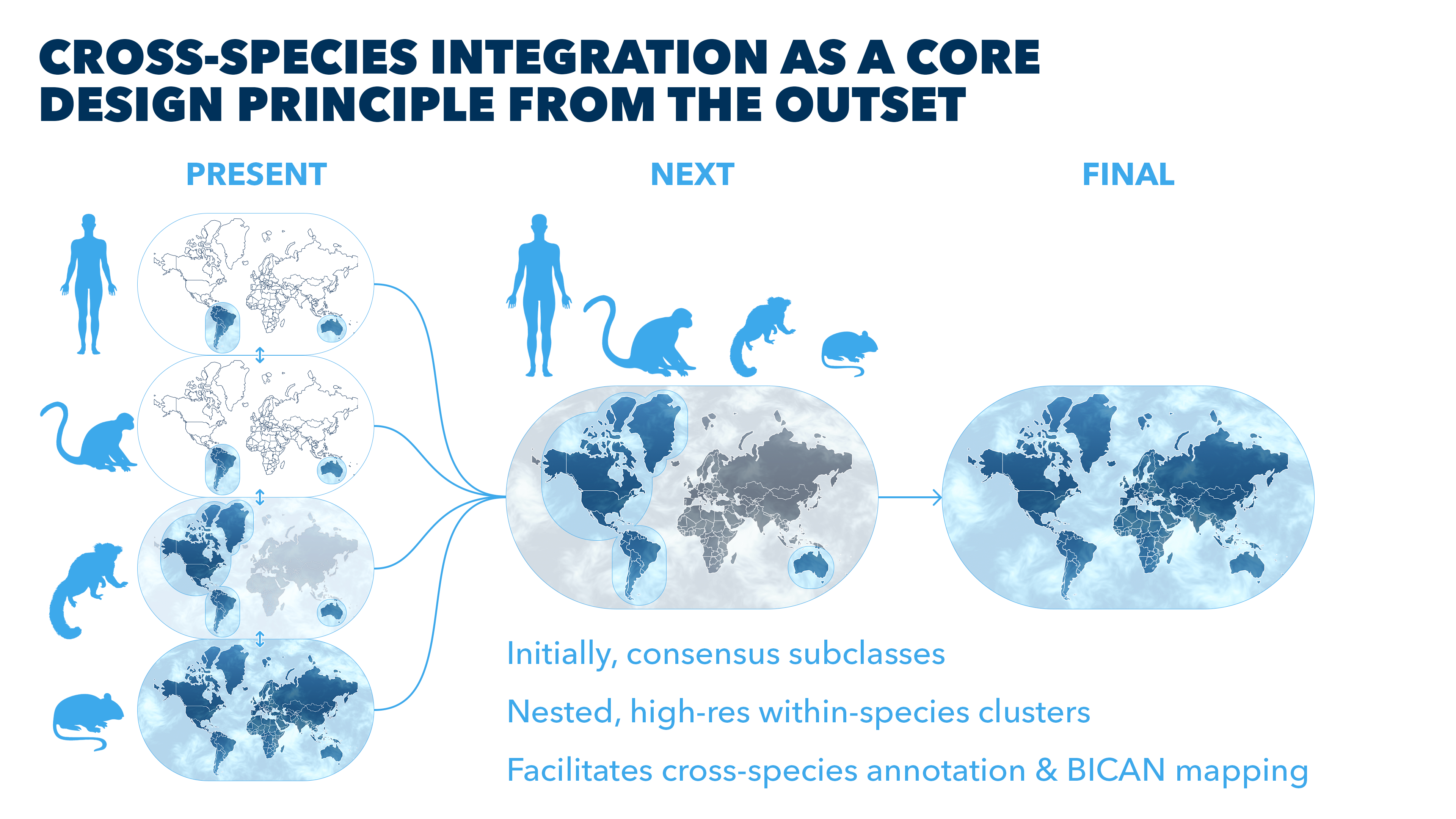
Roadmap toward the cross-species whole-brain reference
Funded through the NIH BRAIN Initiative Cell Atlas Network (BICAN) and building on prior whole brain mouse cell atlas and extensive piloting on human and nonhuman primate (NHP), the HMBA takes a comparative strategy in human, macaque and marmoset that maximizes the strengths of NHP to understand human brain cellular organization and specialization and its relationship to fMRI-based functional architecture. The reference created by HMBA intends to:
- Map genomically defined cell types across the entire human and NHP brain
- Align cell types across species from mouse, marmoset, macaque and human, to produce a cross species
- Map cell types to structural and functional architecture
- Map spatial distributions of genomically defined cell types
- Characterize morphoelectric properties of genomically defined cell types
- Understand the robustness/variability of cell types across individuals and mammalian species
The outline of the cell type world is already in place, with Siletti et al 2023 describing broad cell types across the whole brain and Yao et al 2023 characterizing the entire adult mouse brain in high-resolution. Now, the first release of HMBA profiles and characterizes the basal ganglia (BG) in high-resolution across human, macaque, and marmoset adding more species to the world map of cell types, with varying resolution across them.
The Basal Ganglia are evolutionarily conserved subcortical brain structures that are essential for motor control, learning, emotion, and cognition and are implicated in neurological and psychiatric disorders. HMBA BG is a multiomic consensus atlas comprising over two million nuclei from human, macaque, and marmoset spanning eight major structures of the BG circuit. Integration of cross-species transcriptomics, epigenomics, spatial transcriptomics, morphology and electrophysiology enabled multimodal characterization of BG cell types and reveals both conserved and divergent features. This comprehensive, multiomic,cross-species BG atlas serves as a foundational resource for understanding circuit evolution, function, and disease.
In a subsequent release, HMBA intends to further integrate subcortical insights, particularly from the marmoset, and to harmonize deeply with the whole brain adult mouse taxonomies from the BRAIN Initiative. Ultimately HMBA intends to fully harmonize these four mammalian species with high-resolution, whole-brain, multi-modal characterization of cell types anatomy - the complete map of the world to be the community's standard reference.
About HMBA
The Human and Mammalian Brain Atlas (HMBA) is a major atlas of the BRAIN Initiative Cell Atlas Network (BICAN) that proposes to establish a comprehensive, highly granular cell atlas in complete adult human, macaque, and marmoset brains that links brain structure, function and cellular architecture. In addition to a comprehensive survey of the adult human a strength of the HMBA is the use of NHPs as model organisms to better understand human brain organization and disease. Creating a new foundational cell atlas that is relevant to both the cellular and systems neuroscience communities require much more than simply running various single-cell assays on sets of imprecisely mapped, retrospectively banked brain tissues to create a single cell genomics-based clustering.