
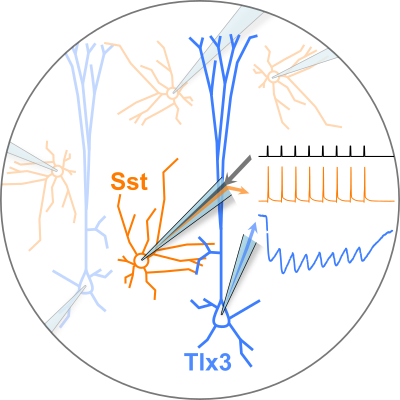
The Allen Institute for Brain Science aims to further our understanding of neuronal cell types by describing the patterns of connectivity among them and characterizing their synaptic signaling. The Synaptic Physiology project advances this goal by examining the intralaminar connectivity between neuronal subclasses in human and mouse cortex via in vitro electrophysiology.
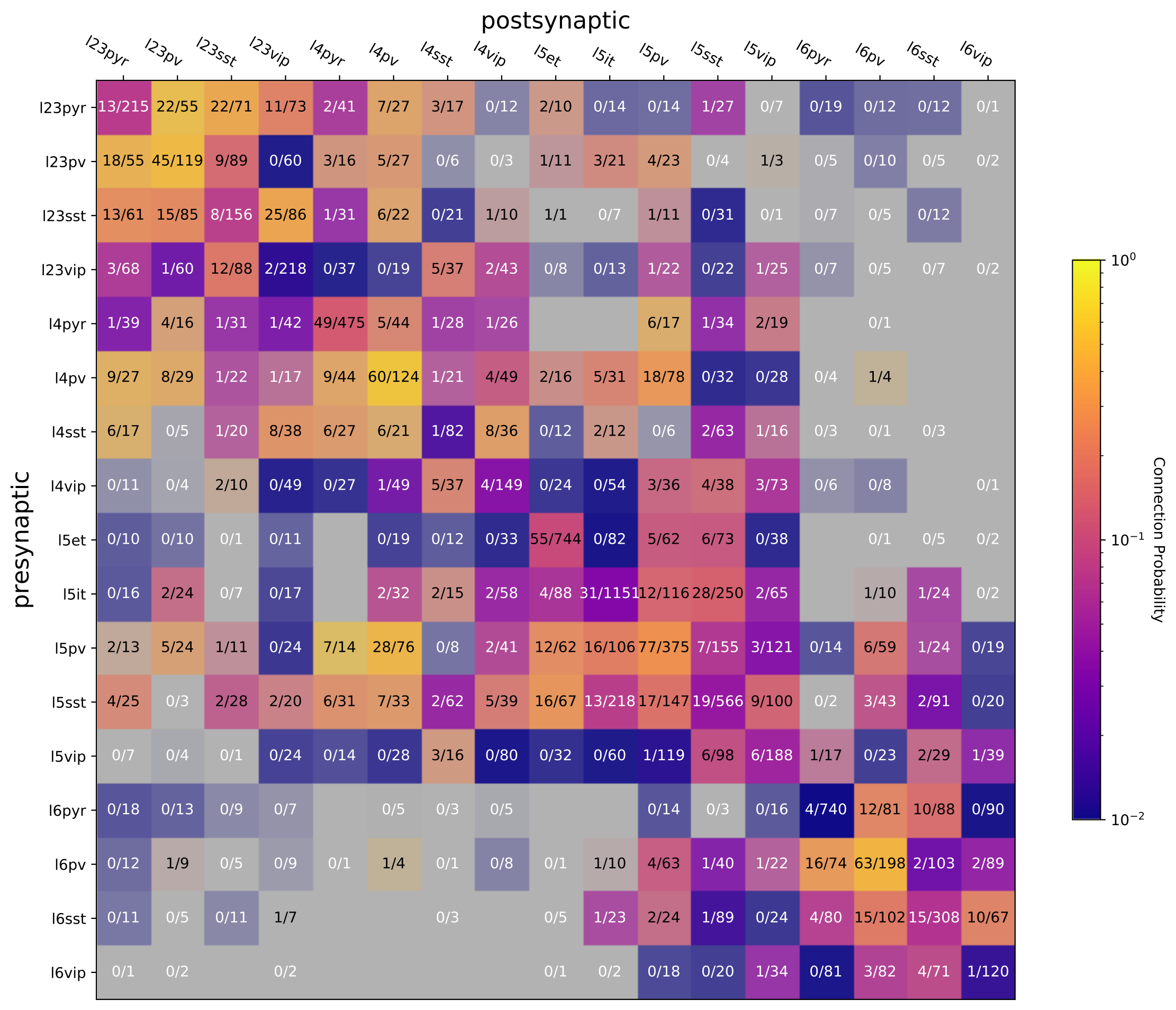
In the mouse dataset, over 23,000 cell pairs were "probed" for connectivity, meaning that we attempted to detect a synaptic connection from one cell onto another. Of those probed pairs, about 1800 had detectable connections, giving an overall probability of about 8% that two cells are synaptically connected. However, this estimate is derived from a mixture of cell types that are known to have different levels of connectivity. We can begin to group connections together based on their pre- and postsynaptic cell subclasses. In the mouse tissue, this usually means classifying cells based on expression of transgenic reporters as well as their laminar position in cortex.
The figure to the left provides an overview of the range of cell subclasses that were tested in the mouse dataset: a focus on intralaminar connectivity between five different excitatory cell subclasses and three inhibitory interneuron subclasses.
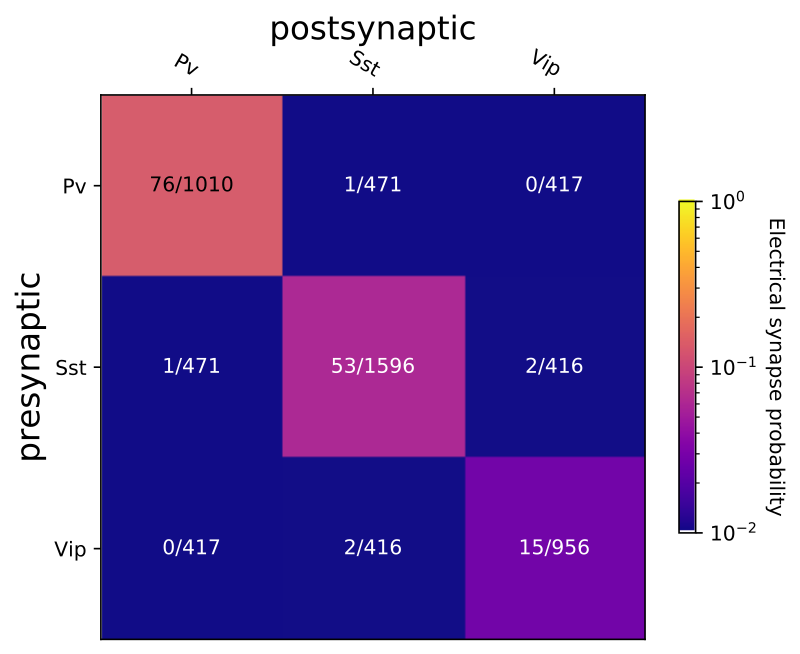
The three inhibitory interneuron subclasses, Pv, Sst, and Vip were also electrically coupled by gap junctions.
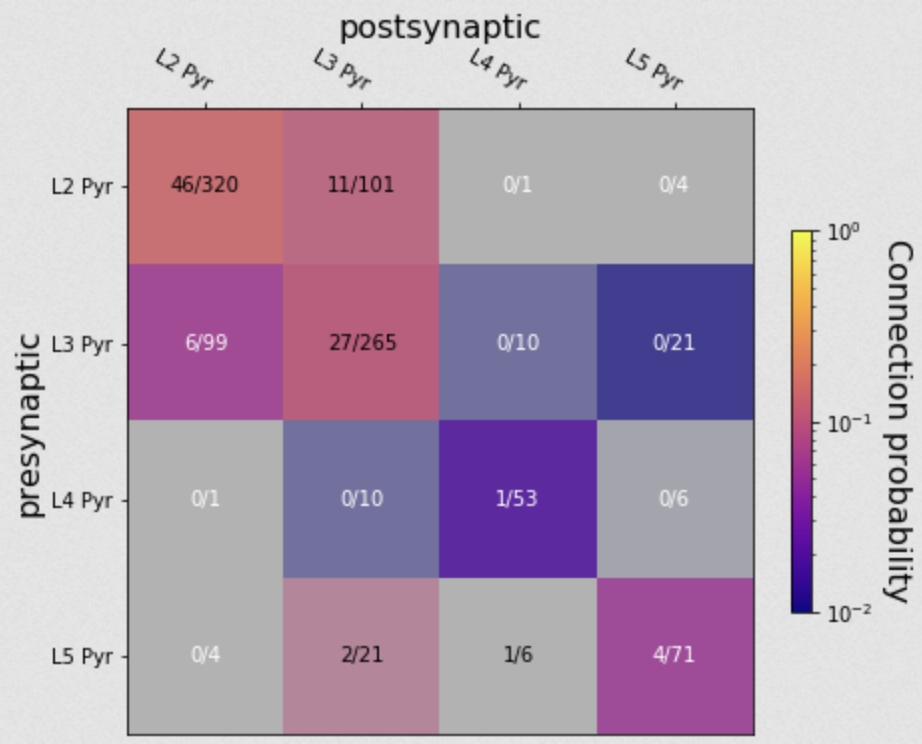
In human tissue, we do not have access to transgenic reporters as a means of classifying cells. Instead, we rely more heavily on morphological features to separate excitatory from inhibitory classes, and laminar position to separate subclasses.
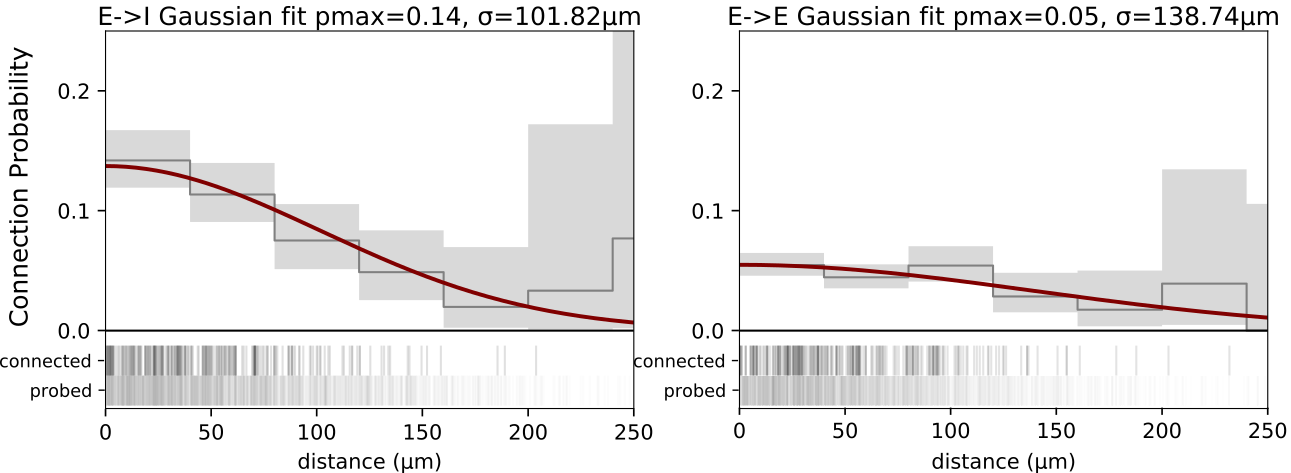
In addition to cell type, the distance between two cell bodies is strongly correlated with the probability of connectivity. Connection distances are not carefully controlled in this dataset, so any sampling bias can cause misleading differences in connectivity to appear. To account for this bias, we can look at the relationship between connection probability and intersomatic distance and model it with a Gaussian.
We see that there is a steep relationship between intersomatic distance and the probability of connectivity. This relationship is further influenced by cell class, in this case impacting E->I connections more so than E->E connections. We can generate a model of this relationship to calculate an unbiased estimate of connectivity in a cell type specific way.
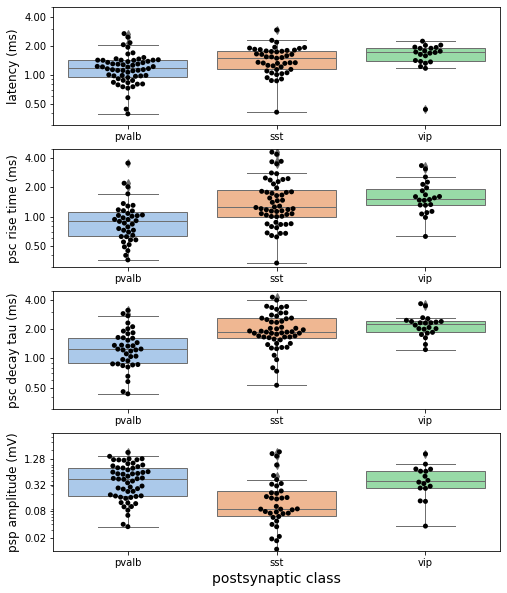
For each connection identified in our dataset we measure the latency, strength, and rise / decay kinetics. These values are mainly derived from curve fits to the averaged responses from both current and voltage clamp recordings. With these results one can make comparisons of kinetic parameters across cell types or construct models with biophysically realistic synapse properties.
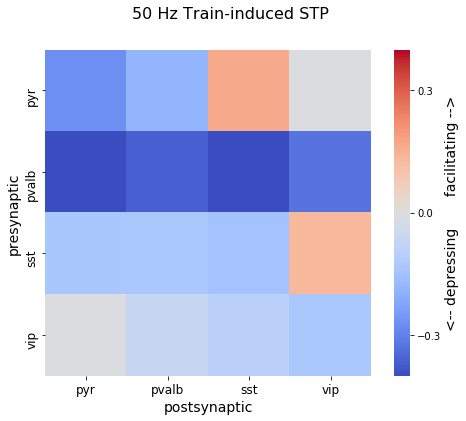
Synapses dynamically vary their strength of signaling over time in a way that is highly stochastic and also depends on the prior history of activity at the synapse. This dynamic nature of synapses increases their computational diversity and is believed to be a major determinant in the behavior of neuronal networks in the brain. The stimuli in our dataset were designed to explore a range of synaptic temporal dynamics.
Per-spike response properties are recorded for every connection in our dataset, but we have also generated three simple metrics that describe some of the most basic features of synaptic short-term plasticity.