As a scientist doing research on the Basal Ganglia, I’m looking for viral genetic tools that allow me to specifically target cell types in the striatum for an upcoming set of experiments.
I use the Genetic Tools Atlas from the Allen Institute to explore whether Allen scientists have publicly shared suitable tools.
I review the provided experiment metadata. I see that at a glance there are several enhancer-adeno-associated viruses (AAVs) targeting the striatum but also ones for many other brain regions.
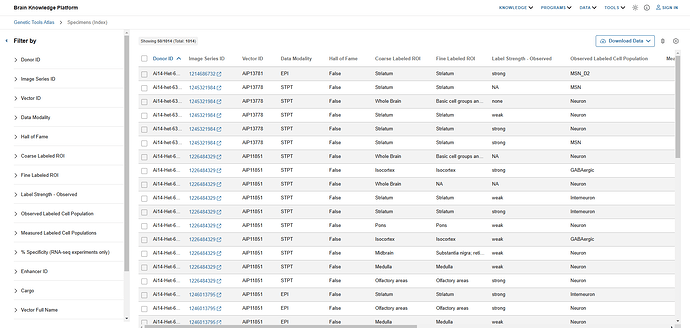
I open the filter panel and find the “Coarse Labeled ROI” filters. I scroll down and select the checkbox next to “Striatum“. I see that there are 372 results that match my query.
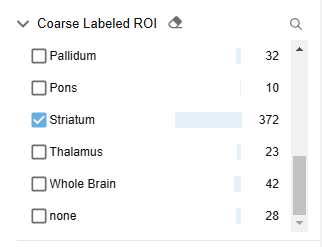
I apply an additional filter to narrow the data to a fine labeled ROI of “Striatum“ and an observed labeled cell population of “Cholinergic“. I’ve narrowed down my search to 36 highly relevant experiments.
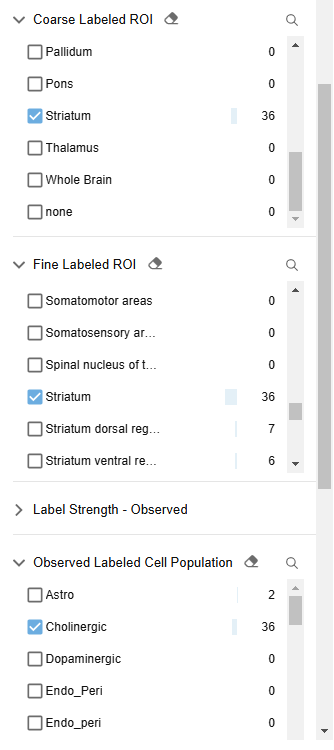
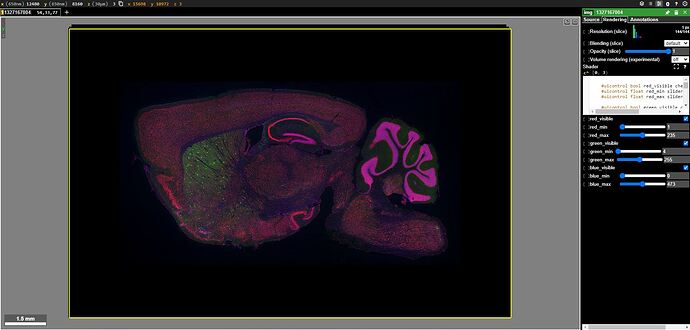
I notice that 2 results use AAVs that were designated as particularly notable, i.e. “Hall of Fame”. I review their EPI & STPT image data. I use Neuroglancer to see how these enhancers are expressed in my regions and cell populations of interest.
Useful Hot-Keys for Neuroglancer:
See the dedicated Neuroglancer documentation for more details.
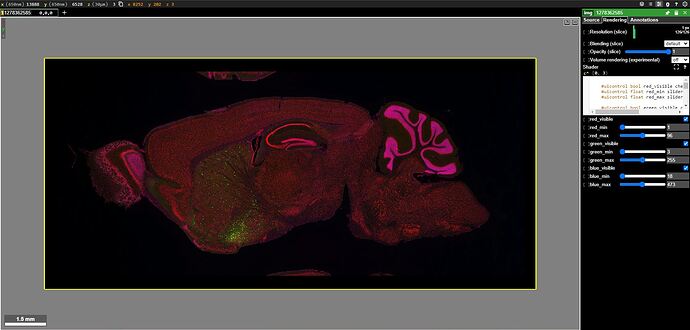
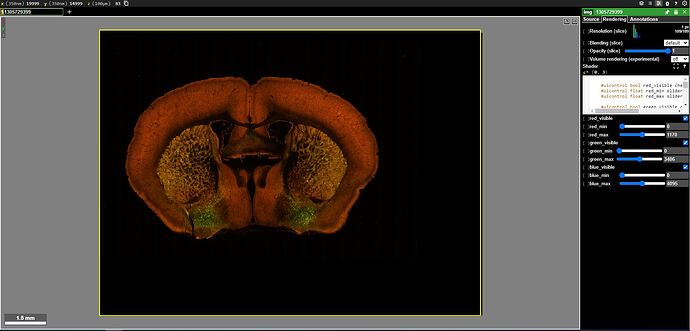
The expression pattern meets my expectations and I decide to use it in future experiments.
I go to http://addgene.org . I type in the Vector ID “AiP14496“ I received from Genetic Tools Atlas and hit the Search button.
The results return one relevant enhancer:

I click into the enhancer entry to access further details and ordering information.

I explore the other results and find AiP13038 and its related image data. I use Neuroglancer to see how these enhancers are expressed in my regions and cell populations of interest. I note that its Addgene ID is listed directly in the Genetic Tools Atlas.


I go to addgene.org. I type in the enhancer ID “191720“ I received from Genetic Tools Atlas and hit the Search button.
The results return one relevant enhancer:

I click into the enhancer entry to access further details and ordering information.

Learn to download file manifests for female chimpanzee brain data from the BICAN grant. Access primate neuroscience research datasets.
Learn to access BICAN consortium data at NEMO Archive using BKP file manifests. Efficiently download large-scale brain research datasets.
Take a guided tour of ASAP-PMDB Parkinson's disease data in the ABC Atlas. Explore molecular profiles and pathology across brain regions.