The Allen Developing Mouse Brain Atlas offers colorimetric In Situ Hybridization data at cellular resolution at seven stages of mouse development. Choose among the following options for retrieving the data:

To search for a specific gene by name, symbol, NCBI accession number, or Entrez gene ID, type your query into the text box, making sure the “Gene Search” radio button is selected. As you type, genes that match your search string will be suggested to you. Select an item from the list and click the “Search” button. The results are displayed in a list that shows all genes with expression data that match your query.
Tips for searching:
The following special operators can be used to build queries:
While our automatically-generated semi-quantitative analysis tools (Anatomic Search and Temporal Search) provide an informative global view of gene expression in the developing brain, these tools provide gene expression information primarily about larger brain regions, with limited resolution for precise brain areas. The Annotation Search feature utilizes expert-guided manual annotation performed by Dr. Salvador Martinez of the Neurosciences Institute, Miguel Hernandez University and CSIC in Alicante, Spain.
This feature allows you to search the annotation data for all genes that match specific gene expression criteria. Start by selecting a structure from the developing mouse ontology and then indicate the age(s), the expression intensity(ies), the expression density(ies) and the expression pattern(s). To understand the criteria used to make these manual annotations, please see the Annotation whitepaper in the Documentation tab.
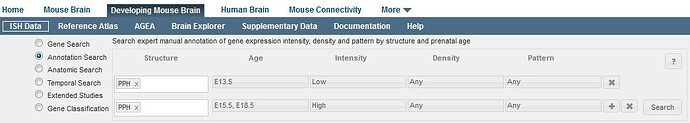
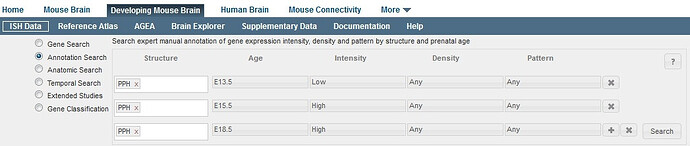
To look for genes that also match additional criteria, click the “+” button to add another row to your search. Remove extra rows by clicking the “x” box. Once you have entered your search criteria, click on the “Search” button. The results are displayed as a list of genes demonstrating gene expression patterns that match your query.
Any genes that meet the criteria in ALL rows of the search will be returned. If two or more specific ages (or intensities, densities or patterns) are selected in a single row, the search will find all results that match any one of the criteria (i.e. an “OR” search).
For example, this query displayed above will return many results because the second line specifies that either the E15.5 or E18.5 timepoints can have high expression in the prepontine hindbrain. The query displayed below looks similar but it is much more selective; only a few genes have high expression in PPH for E15.5 AND E18.5, AND Low expression in E13.5.
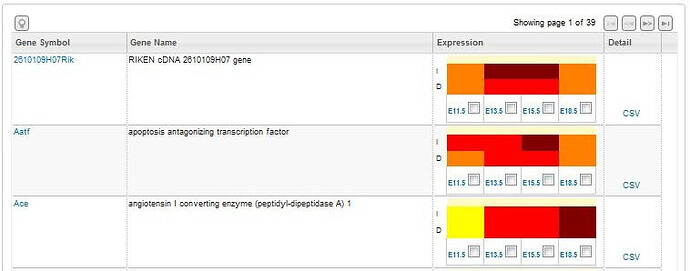
Search returns include a list of experiments that fulfill your search criteria listing the gene symbol, the gene name, an expression summary illustrated in a heatmap format and a link to the .csv annotation document for that gene. The expression summary will include:



Select the “Anatomic Search” radio button to search for genes that are predominantly enriched within the selected brain structure. Results will be returned for the specified age. Experiments are ranked by the specificity of the gene in the brain structure of interest as opposed to adjacent brain regions. The rank is defined as the ratio of sum of expressing pixel intensities in the numerator set (target region) over the sum of expressing pixel intensities in the denominator set (a defined set of non-overlapping structures, see Data Processing Whitepaper under the Documentation tab). Thirteen structures are generally available for search for each age, with the exception of E11.5, for which five structures are available. The results are provided as a list of experiments for the given age and search query.
Please refer to the Informatics Data Processing white paper in the Documentation tab for additional details.
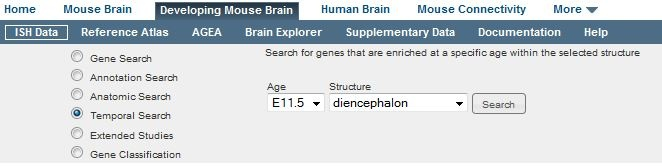
Select the “Temporal Search” radio button to search for genes that are enriched at a specific age within the selected structure. Note that this feature does not search for spatial enrichment; Anatomic Search should be used for that purpose. This search feature compares gene expression in the given structure at the given age and compares it to expression in the same structure at 7 other time points. Results are returned as a list of experiments ranked by their temporal enrichment in descending order, with the most temporally enriched genes first.
Please refer to the Informatics Data Processing white paper in the Documentation tab for additional details.
Once you have found a gene of interest, you can also find genes with a similar expression profile to your gene of interest using the Correlation Search option: once a gene is selected by clicking on the gene name, from the right-hand panel, choose one or more structures from the structure drop-down menu and one or more ages from the ages drop-down menu and then click “Search”. Your search will result in a list of genes with a similar expression pattern to your search criteria.

A select group of genes were sampled deeper and can be viewed using the Extended Studies radio button.
Select “High Temporal Sampling Study” to see ~20 developmental marker genes characterized by ISH in coronal plane with higher temporal resolution, including 8 embryonic developmental stages and 6 early postnatal stages.
Select “Aging Study” to see ~300 genes surveyed in aged brain at 18 and 24 months.
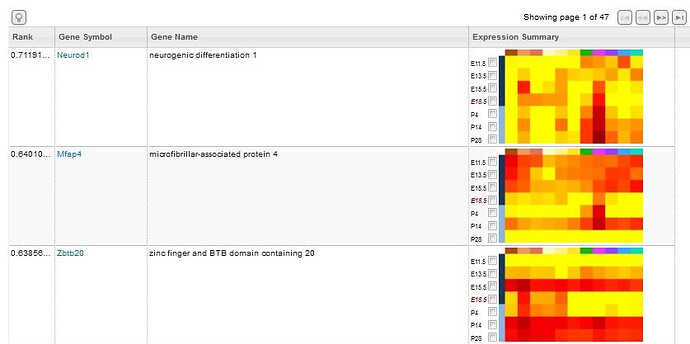
Searches will return a list of genes based on the input search criteria. Each row includes the following information.
Select the experiments to view in greater detail by clicking the checkboxes next to gene and age of interest. Experiments will be saved in your cache until you select the “Clear Selections” button at the bottom of the page. After selecting one or more experiments, click the “View Selections” button.
Once you have selected a gene (by clicking on it’s Gene Name), you can view the rotating 3-D thumbnails in the right hand panel and you can look for genes with a similar expression profile.
The experiment image viewer displays images for each selected experiment in a Zoom and Pan viewer. This viewer makes it easy to compare experiments with each other and with the Reference Atlas.
Multiple image series can be opened on the same page to enable side-by-side comparisons. Arrange the experiments by dragging a viewer by the title bar and dropping in a new location. Add one or more reference atlases by selecting from the “Atlases” drop-down menu in the upper-right hand corner of the window.
If you are viewing more than one experiment, open the configuration options to change the number of columns displayed in the window. The configuration options are accessible by clicking on the button with a “gear” icon to the right of the “Atlases” menu.
The gene symbol is displayed in the title bar along with the Allen Institute image series ID. Additional details are displayed across the top of the viewing area, including the image index, tissue index, and tissue location.
Thumbnails for the entire image series are displayed across the bottom of the viewer in section order. Click a thumbnail to select it for viewing, or use the keyboard to navigate through the set. The current selection is outlined in black.
When you click on the gene symbol, you will be taken to a page that describes the experiments involving this gene in detail. The data returned includes gene detail metadata, the Expression Summary and a list of the experiments performed.
Gene detail metadata includes the name of the gene, any aliases this gene may have, the organism that the experiment was conducted on, the Entre ID and the chromosome location of the gene.

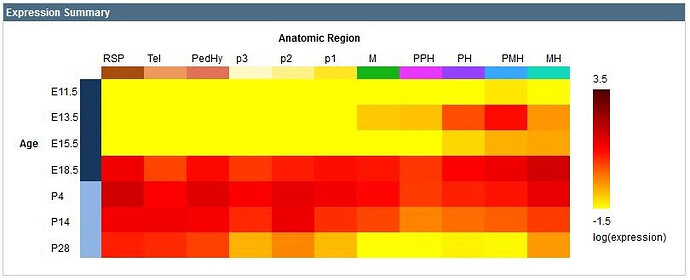
The gene expression summary is represented in heatmap format with the age of the mouse on the vertical axis and the anatomic region on the horizontal axis color-coded to the reference atlas. The heatmap illustrates increasing expression energy ranging from a log value of -1.5 in the yellow to 3.5 in the dark red.
A list of experiments conducted on this gene includes:

Click on one or more experiments to view all the experiments in an interactive environment. This data can also be downloaded as an XML query by clicking on the “This data is also available as XML” link.
When the experiment is selected by clicking on the experimental ID link, a summary of the experimental detail is returned. The data includes 1) Experiment and Specimen metadata and a link to Related Institute Data 2) an interactive 3-dimensional representation of gene expression, 3) an interactive image viewer that displays the images in the experiment 4) a histogram of the expression energy in 11 different structures, and 5) Probe metadata including forward and reverse primers and probe sequence.
Hovering your mouse over the histogram will sync the image in the single image viewer with the section corresponding to that structure and expression.
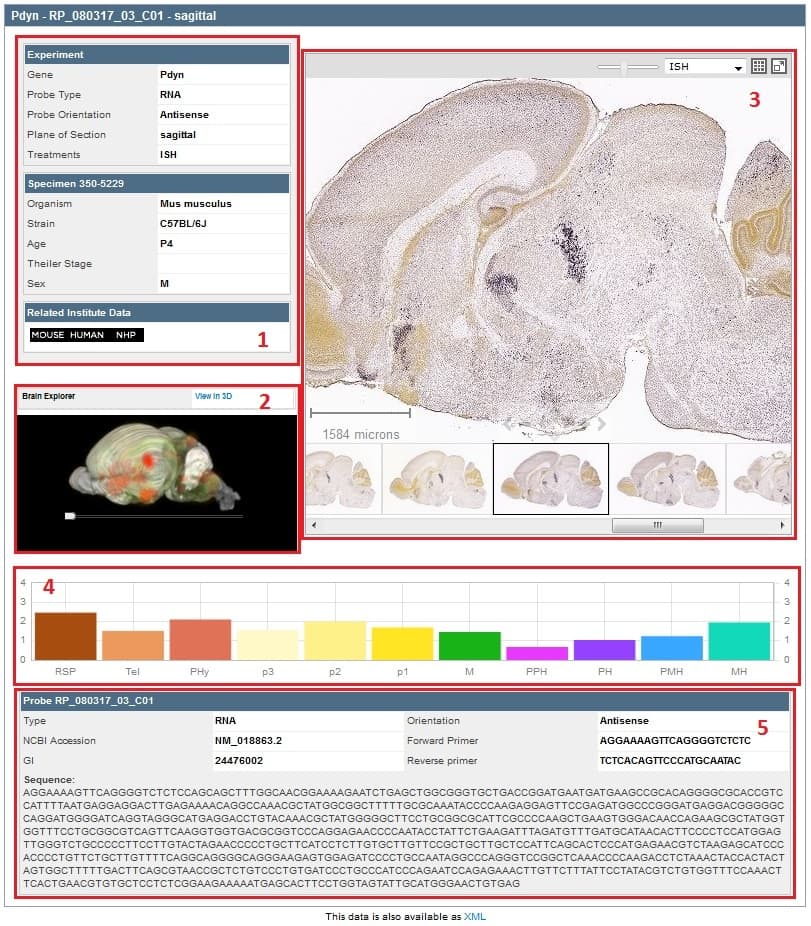
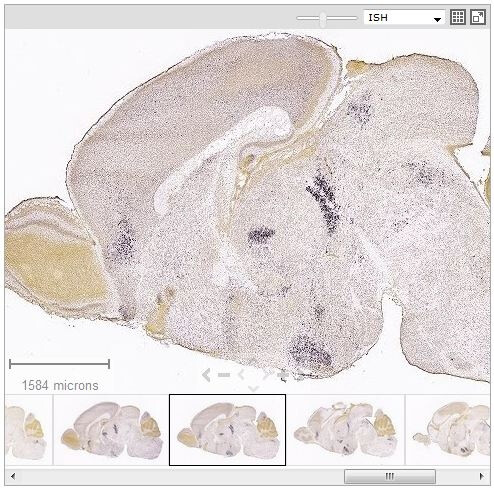
The Zoom and Pan (ZAP) Image Viewer is a powerful tool to navigate and view the images in an experiment. The main part of the viewer is an interactive window where an image can be repositioned by dragging with a mouse. Use the scroll wheel, the on screen navigation tools or the keyboard to zoom in or out.
Select other images in the experiment by clicking on a thumbnail image below the main viewer.
Drag the scale bar with your mouse to the desired location. Click the text with your mouse to toggle between horizontal and vertical.
Use the toolbar to take actions on the image that currently has focus. Toolbar controls include:
Use the keyboard to navigate through the image series and synchronize the viewers on the page. Keyboard commands include:
You can also use the arrow keys to pan the current image.
The expression mask image display highlights those cells that have the highest probability of gene expression using a heat map color scale (from low/blue to high/red).

The Expression Energy was calculated as follows: Within a given area A (voxel or structure), expression energy = (sum of intensity of expressing pixels in A) / (sum of all pixels in A)
The image viewer enables you to view a high resolution image in its own re-sizable window. It is possible to open multiple images by opening multiple windows.
Interacting with the High Resolution Image Viewer is similar to using the ZAP Viewer with some enhanced features accessible from icons in the top right hand of the main image viewer.
Master Allen Brain Explorer desktop application for 3D visualization. Navigate brain atlases, view gene expression, and create custom visualizations.
Learn about API For Allen Developing Mouse Brain Atlas with comprehensive guides and examples from Allen Institute for Brain Science.
Learn about Developmental Anatomic Gene Expression Atlas AGEA with comprehensive guides and examples from Allen Institute for Brain Science.