The BrainSpan Atlas of the Developing Human Brain is a foundational resource for studying transcriptional mechanisms involved in human brain development. The data includes:
From the API, you can:

Download expression values

Query the correlative and differential search services
The Developmental Transcriptome Study is a broad developmental survey of gene expression in specific brain regions using RNA sequencing and exon microarray techniques. The survey profiles up to sixteen targeted cortical and subcortical structures across the full course of human brain development, spanning pre- and postnatal-development in both males and females.
Each sampling site is associated with a Structure. For convenience, surveyed structures are grouped together in the “Developing Human - Transcriptome” StructureSet.
Typically, for each sample four types of expression values are available:
Normalized expression values can be downloaded in several ways:
All experimental data from this study is associated with the “Developing Human Transcriptome” Product. All gene, exon, probeset, donor and sampling site information can be accessed through the API using RMA queries.
Examples:
http://api.brain-map.org/api/v2/data/Donor/query.xml?include=age&criteria=products[id$eq24]
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Specimen, rma::criteria,donor(products[id$eq24]), well_known_files(well_known_file_type[name$eqRNASeqSummarizedToGenes]), rma::include,donor(age)
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Donor, rma::criteria,products[id$eq24],rma::include,age[embryonic$eqtrue]
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Donor, rma::criteria,products[id$eq24],rma::include,age[embryonic$eqfalse]
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Structure, rma::criteria,structure_sets[name$eq’Developing Human - Transcriptome’], rma::options[only$eq’structures.id,structures.acronym,structures.name,structures.color_hex_triplet’]

Normalized expression values can be obtained by specifying:
See the connected service page for definitions of service::dev_human_expression parameters.
|Expression Type | “set” parameter | “probe” object|
|— | — | —|
|RNA-Seq RPKM values summarized to the gene level | rna_seq_genes | Gene (ensembl_id not null )|
|RNA-Seq RPKM values summarized to the exon level | rna_seq_exons | GoExons|
|Exon microarray expression values summarized to the gene level | exon_microarray_genes | Gene (entrez_id not null )|
|Exon microarray expression values summarized to the probeset level | exon_microarray_exons | AffymetriProbsets|
Example:
Download RNA-Seq expression values for samples from donor “h376.VI.52” associated with gene CARTPT
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Donor, rma::criteria,[name$eq’H376.VI.52’], rma::options[only$eq’donors.id’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Gene, rma::criteria,[acronym$eq’CARTPT’][type$eq’EnsemblGene’],organism[name$eq’Homo Sapiens’], rma::options[only$eq’genes.id’]
http://api.brain-map.org/api/v2/data/query.json?criteria=service::dev_human_expression [set$eq’rna_seq_genes’][probes$eq1098278][donors$eq12890]
The output of the service is two top level ordered arrays “probes” and “samples”. For example:
“probes”:[ {“id”:1098278, “name”:“ENSG00000164326”, “gene-id”:9463, “gene-symbol”:“CARTPT”,“gene-name”:“CART prepropeptide”, “entrez-id”:9607,“chromosome”:“5”,“start-position”:“n/a”,“end-position”:“n/a”, “expression_level”:[“5.5566”,“4.9466”,“5.6331”,…]} ], “samples”:[ {“donor”:{“id”:12890,“name”:“H376.VI.52”,“age”:“4 mos”,“color”:“76EB76”}, “structure”:{“id”:10173,“name”:“dorsolateral prefrontal cortex”,“abbreviation”:“DFC”,“color”:“D4B235”}, “top_level_structure”:{“id”:10153,“name”:“neural plate”,“abbreviation”:“NP”,“color”:“D7D8D8”}}, {“donor”:{“id”:12890,“name”:“H376.VI.52”,“age”:“4 mos”,“color”:“76EB76”}, “structure”:{“id”:10185,“name”:“ventrolateral prefrontal cortex”,“abbreviation”:“VFC”,“color”:“C2A335”}, “top_level_structure”:{“id”:10153,“name”:“neural plate”,“abbreviation”:“NP”,“color”:“D7D8D8”}}, {“donor”:{“id”:12890,“name”:“H376.VI.52”,“age”:“4 mos”,“color”:“76EB76”}, “structure”:{“id”:10278,“name”:“anterior (rostral) cingulate (medial prefrontal) cortex”,“abbreviation”:“MFC”,“color”:“E26880”}, “top_level_structure”:{“id”:10153,“name”:“neural plate”,“abbreviation”:“NP”,“color”:“D7D8D8”}}, … ],
Each probe (Gene) contains information about:
Each sample contains information about:
Example:
Download RNA-Seq expression values for samples from donor “h376.VI.52” and all exons associated with gene CARTPT
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Donor, rma::criteria,[name$eq’H376.VI.52’], rma::options[only$eq’donors.id’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::GoExon, rma::criteria,gene_association(gene[acronym$eq’CARTPT’]), rma::options[only$eq’go_exons.id’]
http://api.brain-map.org/api/v2/data/query.json?criteria=service::dev_human_expression [set$eq’rna_seq_exons’][probes$eq279330730,279330740,2793307356][donors$eq12890]
The output of the service is two top level ordered arrays “probes” and “samples”. For example:
“probes”:[ {“id”:279330730, “name”:“ENSG00000164326”, “gene-id”:1098278, “gene-symbol”:“CARTPT”, “gene-name”:“CART prepropeptide”, “entrez-id”:9607,“chromosome”:“5”,“start-position”:71014989,“end-position”:71015279, “expression_level”:[“5.7282”,“4.8102”,“5.7152”, … ]}, {“id”:279330735, “name”:“ENSG00000164326”, “gene-id”:1098278,“gene-symbol”:“CARTPT”, “gene-name”:“CART prepropeptide”, “entrez-id”:9607,“chromosome”:“5”,“start-position”:71015405,“end-position”:71015790, “expression_level”:[“4.4128”,“3.8230”,“4.3135”, … ]}, {“id”:279330740, “name”:“ENSG00000164326”, “gene-id”:1098278,“gene-symbol”:“CARTPT”, “gene-name”:“CART prepropeptide”, “entrez-id”:9607,“chromosome”:“5”,“start-position”:71016334,“end-position”:71016875, “expression_level”:[“5.9593”,“5.4664”,“6.1137”, … ]} ], “samples”:[ {“donor”:{“id”:12890,“name”:“H376.VI.52”,“age”:“4 mos”,“color”:“76EB76”}, “structure”:{“id”:10173,“name”:“dorsolateral prefrontal cortex”,“abbreviation”:“DFC”,“color”:“D4B235”}, “top_level_structure”:{“id”:10153,“name”:“neural plate”,“abbreviation”:“NP”,“color”:“D7D8D8”}}, {“donor”:{“id”:12890,“name”:“H376.VI.52”,“age”:“4 mos”,“color”:“76EB76”}, “structure”:{“id”:10185,“name”:“ventrolateral prefrontal cortex”,“abbreviation”:“VFC”,“color”:“C2A335”}, “top_level_structure”:{“id”:10153,“name”:“neural plate”,“abbreviation”:“NP”,“color”:“D7D8D8”}}, {“donor”:{“id”:12890,“name”:“H376.VI.52”,“age”:“4 mos”,“color”:“76EB76”}, “structure”:{“id”:10278,“name”:“anterior (rostral) cingulate (medial prefrontal) cortex”,“abbreviation”:“MFC”,“color”:“E26880”}, “top_level_structure”:{“id”:10153,“name”:“neural plate”,“abbreviation”:“NP”,“color”:“D7D8D8”}}, … ],
Each probe (GoExon) contains information about:
Each sample contains information about:
Example:
Download exon microarray expression values for samples from donor “h376.VI.50” associated with gene CARTPT
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Donor, rma::criteria,[name$eq’H376.VI.50’], rma::options[only$eq’donors.id’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Gene, rma::criteria,[acronym$eq’CARTPT’][type$eq’NcbiGene’],organism[name$eq’Homo Sapiens’], rma::options[only$eq’genes.id’]
http://api.brain-map.org/api/v2/data/query.json?criteria=service::dev_human_expression [set$eq’exon_microarray_genes’][probes$eq9463][donors$eq12296]
The output of the service is two top level ordered arrays “probes” and “samples”. For example:
“probes”:[ {“id”:9463, “name”:“ENSG00000164326”, “gene-id”:9463, “gene-symbol”:“CARTPT”, “gene-name”:“CART prepropeptide”, “entrez-id”:9607,“chromosome”:“5”,“start-position”:“n/a”,“end-position”:“n/a”, “expression_level”:[“8.1073”,“5.9564”,“6.3102”,“7.0532”,“9.6157”,“7.1290”,“5.1168”,“4.4212”,“4.2779”], “z-score”:[“1.3547”,“0.0538”,“0.2678”,“0.7172”,“2.2670”,“0.7630”,“-0.4541”,“-0.8748”,“-0.9615”]} ], “samples”:[ {“donor”:{“id”:12296,“name”:“H376.VI.50”,“age”:“4 mos”,“color”:“80FF80”}, “structure”:{“id”:10278,“name”:“anterior (rostral) cingulate (medial prefrontal) cortex”,“abbreviation”:“MFC”,“color”:“E26880”}, “top_level_structure”:{“id”:10153,“name”:“neural plate”,“abbreviation”:“NP”,“color”:“D7D8D8”}}, {“donor”:{“id”:12296,“name”:“H376.VI.50”,“age”:“4 mos”,“color”:“80FF80”}, “structure”:{“id”:10243,“name”:“posterior (caudal) superior temporal cortex (area 22c)”,“abbreviation”:“STC”,“color”:“D670A0”}, “top_level_structure”:{“id”:10153,“name”:“neural plate”,“abbreviation”:“NP”,“color”:“D7D8D8”}}, … ]
Each probe (Gene) contains information about:
Each sample contains information about:
Download exon microarray expression values for samples from donor “h376.VI.50” for all probesets associated with gene CARTPT
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Donor, rma::criteria,[name$eq’H376.VI.50’], rma::options[only$eq’donors.id’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::AffymetrixProbeset, rma::criteria,gene_association(gene[acronym$eq’CARTPT’](organism[name$eq’Homo Sapiens’])), rma::options[only$eq’affymetrix_probesets.id’]
http://api.brain-map.org/api/v2/data/query.json?criteria=service::dev_human_expression [set$eq’exon_microarray_exons’][probes$eq280550735,280550750,280550740,280550730,280550745][donors$eq12296]
The output of the service is two top level ordered arrays “probes” and “samples”. For example:
“probes”:[ {“id”:280550735, “name”:“ENSG00000164326”, “gene-id”:9463, “gene-symbol”:“CARTPT”, “gene-name”:“CART prepropeptide”, “entrez-id”:9607,“chromosome”:“5”,“start-position”:71015125,“end-position”:71015265, “expression_level”:[“8.1073”,“6.4681”,“6.3102”,“7.0532”,“9.4350”,“7.1290”,“5.1560”,“5.0550”,“4.9879”], “z-score”:[“1.4783”,“0.0367”,“-0.1021”,“0.5513”,“2.6458”,“0.6179”,“-1.1171”,“-1.2059”,“-1.2649”]}, {“id”:280550750, “name”:“ENSG00000164326”, “gene-id”:9463, “gene-symbol”:“CARTPT”, “gene-name”:“CART prepropeptide”, “entrez-id”:9607,“chromosome”:“5”,“start-position”:71016613,“end-position”:71016799, “expression_level”:[“10.1951”,“8.8021”,“7.7846”,“9.4317”,“11.8110”,“10.0210”,“6.1850”,“5.6075”,“4.5526”], “z-score”:[“1.2342”,“0.6566”,“0.2348”,“0.9177”,“1.9041”,“1.1620”,“-0.4284”,“-0.6678”,“-1.1052”]}, ], “samples”:[ {“donor”:{“id”:12296,“name”:“H376.VI.50”,“age”:“4 mos”,“color”:“80FF80”}, “structure”:{“id”:10278,“name”:“anterior (rostral) cingulate (medial prefrontal) cortex”,“abbreviation”:“MFC”,“color”:“E26880”}, “top_level_structure”:{“id”:10153,“name”:“neural plate”,“abbreviation”:“NP”,“color”:“D7D8D8”}}, {“donor”:{“id”:12296,“name”:“H376.VI.50”,“age”:“4 mos”,“color”:“80FF80”}, “structure”:{“id”:10243,“name”:“posterior (caudal) superior temporal cortex (area 22c)”,“abbreviation”:“STC”,“color”:“D670A0”}, “top_level_structure”:{“id”:10153,“name”:“neural plate”,“abbreviation”:“NP”,“color”:“D7D8D8”}}, … ]
Each probe (Gene) contains information about:
Each sample contains information about:
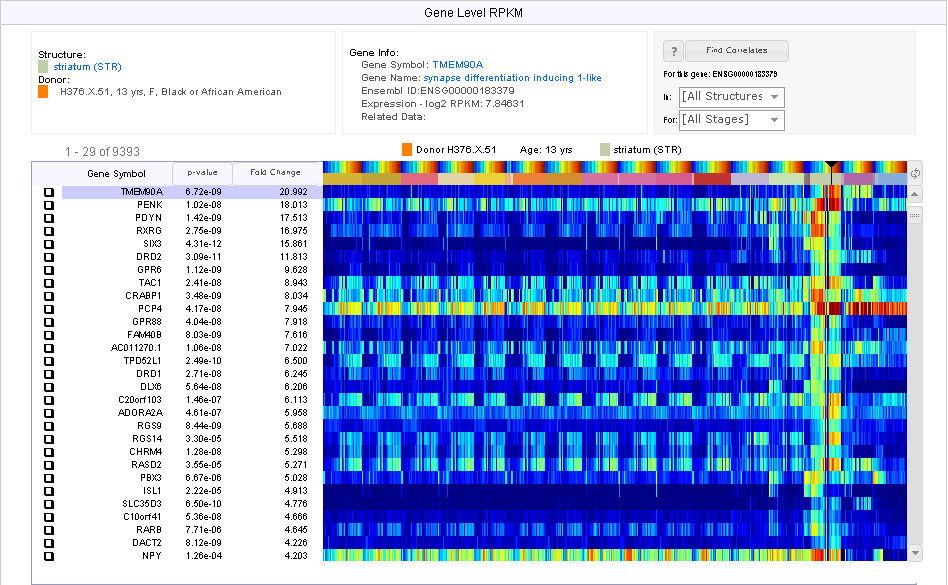
Differential search find “probes” that show the greatest difference in expression values between two sets (target and contrast) of user-defined structures. For each probe, a 2-sample t-test is performed followed by Benjamini and Hochberg false discovery rate correction. The null hypothesis is that the average expression level of samples in the contrast set of structures is greater than or equal to the average expression level of samples in the target set of structures. A statistically significant result (p-value less than user-defined threshold) allows us to reject the null hypothesis and conclude that the average expression level of samples in the target set of structures is greater than the average expression level of samples in the contrast set of structures. Resulting p-values are sorted in ascending order. Search results can also be sorted by fold-change (log ratio of expression) in descending order.

The differential search function can be accessed through the Web application or using the API.
See the connected service page for definitions of service::dev_human_differential parameters.
Example:
Differential search for genes with higher expression in “striatum” than in the whole “neural plate” over all donors for each of the four types of expression data.
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Structure, rma::criteria,[name$il’striatum’],ontology[name$eq’Developing Human Brain Atlas’], rma::options[only$eq’structures.id’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Structure, rma::criteria,[name$il’neural plate’],ontology[name$eq’Developing Human Brain Atlas’], rma::options[only$eq’structures.id’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=service::dev_human_differential [set$eq’rna_seq_genes’][structures1$eq10153][structures2$eq10333][sort_by$eq’fold-change’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=service::dev_human_differential [set$eq’rna_seq_exons’][structures1$eq10153][structures2$eq10333][sort_by$eq’fold-change’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=service::dev_human_differential [set$eq’exon_microarray_genes’][structures1$eq10153][structures2$eq10333][sort_by$eq’fold-change’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=service::dev_human_differential [set$eq’exon_microarray_exons’][structures1$eq10153][structures2$eq10333][sort_by$eq’fold-change’]
Example:
Differential search for genes with higher expression at 8-9 pcw than at 10-12 pcw over the whole “neural plate” for each of the four types of expression data.
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Structure, rma::criteria,[name$il’neural plate’],ontology[name$eq’Developing Human Brain Atlas’], rma::options[only$eq’structures.id’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Donor, rma::criteria,products[name$eq’Developing Human Transcriptome’],age[embryonic$eqtrue][days$ge56][days$le63], rma::options[only$eq’donors.id’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=model::Donor, rma::criteria,products[name$eq’Developing Human Transcriptome’],age[embryonic$eqtrue][days$ge70][days$le84], rma::options[only$eq’donors.id’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=service::dev_human_differential [set$eq’rna_seq_genes’][donors1$eq12835,12960,13060][donors2$eq12833,13058] [structures1$eq10153][structures2$eq10153][sort_by$eq’fold-change’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=service::dev_human_differential [set$eq’rna_seq_exons’][donors1$eq12835,12960,13060][donors2$eq12833,13058] [structures1$eq10153][structures2$eq10153][sort_by$eq’fold-change’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=service::dev_human_differential [set$eq’exon_microarray_genes’][donors1$eq12835,12960,13060][donors2$eq12833,13058] [structures1$eq10153][structures2$eq10153][sort_by$eq’fold-change’]
http://api.brain-map.org/api/v2/data/query.xml?criteria=service::dev_human_differential [set$eq’exon_microarray_exons’][donors1$eq12835,12960,13060][donors2$eq12833,13058] [structures1$eq10153][structures2$eq10153][sort_by$eq’fold-change’]
Correlative search finds “probes” with expression profile similar that of a to a selected seed “probe” over all samples within a user-specified structure and for user-specified donors. Pearson’s correlation coefficient is computed for all probes and the results ranked in descending order.

The correlative search function can be accessed through the Web application or using the API.
See the connected service page for definitions of service::dev_human_correlation parameters.
Example:
Correlative search for “probes” with similar expression to PVALB over the whole brain
http://api.brain-map.org/api/v2/data/query.xml?criteria= service::dev_human_correlation[set$eqrna_seq_genes][probes$eq1089164][structures$eq10153]
http://api.brain-map.org/api/v2/data/query.xml?criteria= service::dev_human_correlation[set$eqrna_seq_exons][probes$eq278724739][structures$eq10153]
http://api.brain-map.org/api/v2/data/query.xml?criteria= service::dev_human_correlation[set$eqexon_microarray_genes][probes$eq5784][structures$eq10153]
http://api.brain-map.org/api/v2/data/query.xml?criteria= service::dev_human_correlation[set$eqexon_microarray_exons][probes$eq281337753][structures$eq10153]
Learn about Brainspan In Situ Hybridization ISH Data with comprehensive guides and examples from Allen Institute for Brain Science.
Learn about Brainspan Searching The Developmental Transcriptome with comprehensive guides and examples from Allen Institute for Brain Science.
Learn about Brainspan Downloading Data with comprehensive guides and examples from Allen Institute for Brain Science.