The Allen Cell Types Database provides multimodal single cell characterization data to enable data-driven approaches to cell type classification.
From the API, you can:

Download electrophysiology data in Neurodata Without Borders (NWB) format

Download computed electrophysiology features

Download cell morphology images

Download morphological reconstructions in SWC format

Download computed morphology features

Download neuronal models trained on this data set
This document provides a brief overview of the data, database organization and example queries. API database object names are in camel case. See the main API Documentation for more information on data models and query syntax.
The accompanying Allen Software Development Kit (SDK) provides python code for accessing electrophysiology data (NWB files) for all cells and morphological reconstructions (SWC files) for a subset of cells. The Allen SDK also provides sample code demonstrating how to download neuronal model parameters and run your own simulations using stimuli for the experiments or custom current injections.
All data used in the web application is available in the ApiCellTypesSpecimenDetail table. Data includes structure, cortical layer, dendrite type (spiny, aspiny, sparsely spiny, n/a), apical dendrite status (intact, truncated, n/a), and others. Mouse-specific records include the transgenic line name and reporter status. Mouse-specific records include disease condition (epilepsy, tumor, none) and years of seizure history.
See whitepapers for detailed experimental and annotation information.
From the API, detailed information about cells can be obtained using RMA queries.
Examples:
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::ApiCellTypesSpecimenDetail
,rma::options[num_rows$eqall]
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::ApiCellTypesSpecimenDetail
,rma::criteria,[tag__dendrite_type$eq'spiny']
,rma::options[num_rows$eqall]
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::ApiCellTypesSpecimenDetail
,rma::criteria,[structure__layer$eq'4']
,rma::options[num_rows$eqall]
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::ApiCellTypesSpecimenDetail
,rma::criteria,[donor__species$il'homo sapiens']
,rma::options[num_rows$eqall]
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::ApiCellTypesSpecimenDetail
,rma::criteria,[donor__disease_state$il'epilepsy']
,rma::options[num_rows$eqall]
All cells in the Allen Cell Types Database have electrophysiological recordings of responses to stimuli from a common set of current injection protocols. See the electrophysiology overview whitepaperfor details on specimen selection, tissue processing, recording, and quality control.
The Cell Types Database categorizes detailed stimulus protocols into set of high level descriptions:
Stimulus sweeps that pass quality control standards are available for download as Neurodata Without Borders (NWB) files. To find the NWB download link for Rorb cell specimen 320654829, use this query:
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::Specimen
,rma::criteria,[id$eq320654829]
,rma::include,ephys_result(well_known_files(well_known_file_type[name$eqNWBDownload]))
The Allen SDK provides a simple Python module to support downloading metadata and NWB files for cells in the Cell Types Database. Please see the Data API Client documentation page to see an example.
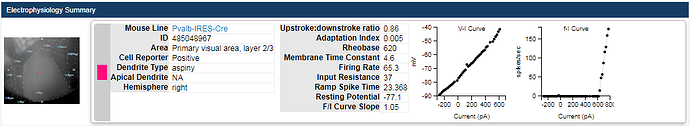
A standard set of electrophysiological features are automatically computed from the recorded responses of each cell. A subset of those features are displayed at the top of the electrophysiology page for a cell:
See the electrophysiology overview whitepaper for a complete list of computed features and their interpretations.
Use this query to download the features computed for Scnn1a cell specimen 467703703:
The Allen Cell Types Database contains morphological reconstructions generated from bright-field images of biocytin-stained cells. Reconstructions are generated by manually curating the results of an automated segmentation algorithm. See the morphology technical whitepaper for more details.
A standard set of morphological features were computed for all reconstructed cells. A subset of those features are displayed at the top of the cell-specific morphology page:
See the morphology technical morphology technical whitepaper for a complete list of computed morphological features.
The API provides programmatic access to the microscopy images used for reconstruction, axis-oriented projections of those images, and morphological reconstructions. A cell can have up to four axis-oriented projections of the images used for reconstruction:
The reconstruction images display a dark, biocytin-filled cell on a light background. The maximum intensity projections are constructed from inverted and contrast-enhanced versions of the morphology images, resulting in a light cell on a dark background.
Examples:
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::ProjectionImage
,rma::criteria,[specimen_id$eq313862022]
http://api.brain-map.org/api/v2/section_image_download/323637357
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::SubImage
,rma::criteria,data_set[specimen_id$eq313862022]
http://api.brain-map.org/api/v2/section_image_download/321549675
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::ApiCellTypesSpecimenDetail
,rma::criteria,[nr__reconstruction_type$nenull]
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::NeuronReconstruction
,rma::criteria,[specimen_id$eq313862306],rma::include,well_known_files
http://api.brain-map.org/api/v2/well_known_file_download/491119517
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::NeuronReconstruction
,rma::criteria,[specimen_id$eq313862022]
The Allen Cell Types Database contains two types of neuronal models: perisomatic biophysical models and generalized leaky integrate-and-fire (GLIF) models. These models attempt to mathematically reproduce a cell’s recorded response to a current injection. The perisomatic biophysical models take into account dendritic morphological structure, whereas GLIF models are simple point neuron models which represent the neuron as a single compartment.
There are five levels of GLIF models with increasing levels of complexity. The most basic model is a simple leaky integrate-and-fire equation. More advanced GLIFs attempt to model variable spike threshold, afterspike currents, and threshold adaptation.
See the perisomatic biophysical and GLIF technical whitepapers for more details on how these models were created.
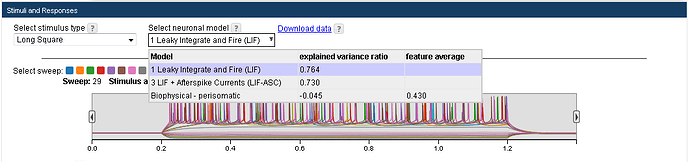
A cell’s electrophysiology page displays all available models. Choose a model to see its simulated response to all stimuli presented to the cell. If the required sweeps are available, two model evaluation metrics are computed per model:
After selecting a model for display, the models ID number and a link for downloading necessary to run the model are available. For example, the link to download model 566296565 (a LIF model for Scnn1a cell 467703703) looks like this:
http://api.brain-map.org/neuronal_model/download/566296565
All models in the Allen Cell Types Database are available for download and local execution via the Allen Software Development Kit (SDK). The biophysical models require NEURON to be run, which the SDK helps to configure. The GLIF simulation module comes as part of the Allen SDK. Please visit the Allen SDK page for more details.
Examples:
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::ApiCellTypesSpecimenDetail
,rma::criteria,[m__biophys_perisomatic$gt0]
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::ApiCellTypesSpecimenDetail
,rma::criteria,[m__glif$gt0]
http://api.brain-map.org/api/v2/data/query.xml?criteria=
model::NeuronalModel
,rma::critera,[specimen_id$eq469803127],neuronal_model_template[name$il'*LIF*']
http://api.brain-map.org/neuronal_model/download/566302806
http://alleninstitute.github.io/AllenSDK/glif_models.html#downloading-glif-models
http://celltypes.brain-map.org/mouse/experiment/electrophysiology/469803127Learn about Cell Types Database Transcriptomics with comprehensive guides and examples from Allen Institute for Brain Science.
Learn about Cell Types Database Physiology And Morphology with comprehensive guides and examples from Allen Institute for Brain Science.
Learn about Documentation Cell Types Database with comprehensive guides and examples from Allen Institute for Brain Science.