Under the ISH tab, there are several ways to search the database.
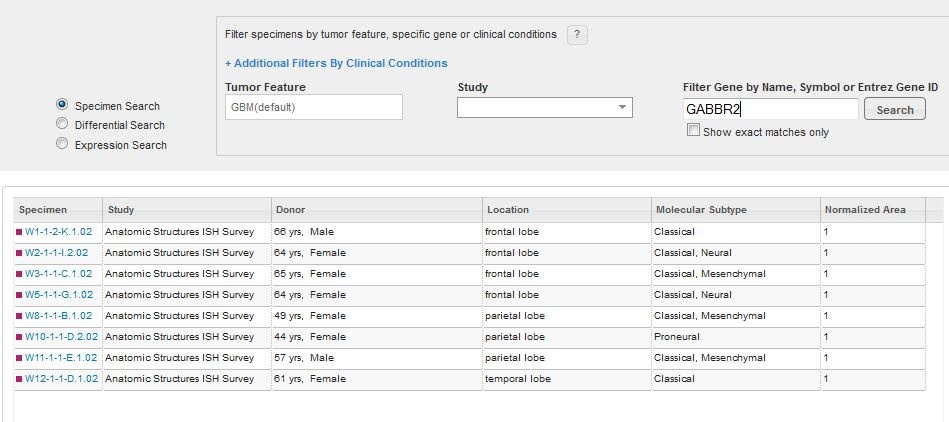
To search for a specific glioblastoma specimen type, filter your search criteria by a specific gene, tumor feature, study or specific clinical conditions.

To refine your search by specific clinical conditions, click the “+” next to “Additional Filters By Clinical Conditions”.
If you are unsure of where to begin your search, you can browse by the tumor information where the columns are described below.
The genes chosen for this project were selected based upon current knowledge of GBM biology and what was learned from each of the individual Ivy GAP Studies. To see a list of genes for each study, please see the Gene Lists whitepaper in Documentation. Typing in the gene search text box will bring up a list of suggested genes. If your gene of interest was not assayed in this project, no results will be returned. Once you have chosen a gene, either by selecting a gene from the suggested list, or hitting “Enter” after typing in the name, symbol or ID, a list of specimen blocks that contain search results fulfilling your criteria will open.
Each of the tumor structural features was identified and labeled in ~12,000 H&E histological images using a semi-automated annotation application based on advanced statistical machine learning algorithms, and can be searched and visualized in the Ivy GAP web application.
Tumor features are described using a specific ontology. The Ivy GAP ontology is a hierarchical organization of glioblastoma anatomic structural features and associated transcriptomes from RNA-Seq samples that were identified by reference histology or reference gene expression patterns. It does not reflect ontological origins of the structural features; it is simply a representation of relationships among the glioblastoma features and gene expression patterns associated with putative cancer stem cell clusters that were identified in the project.
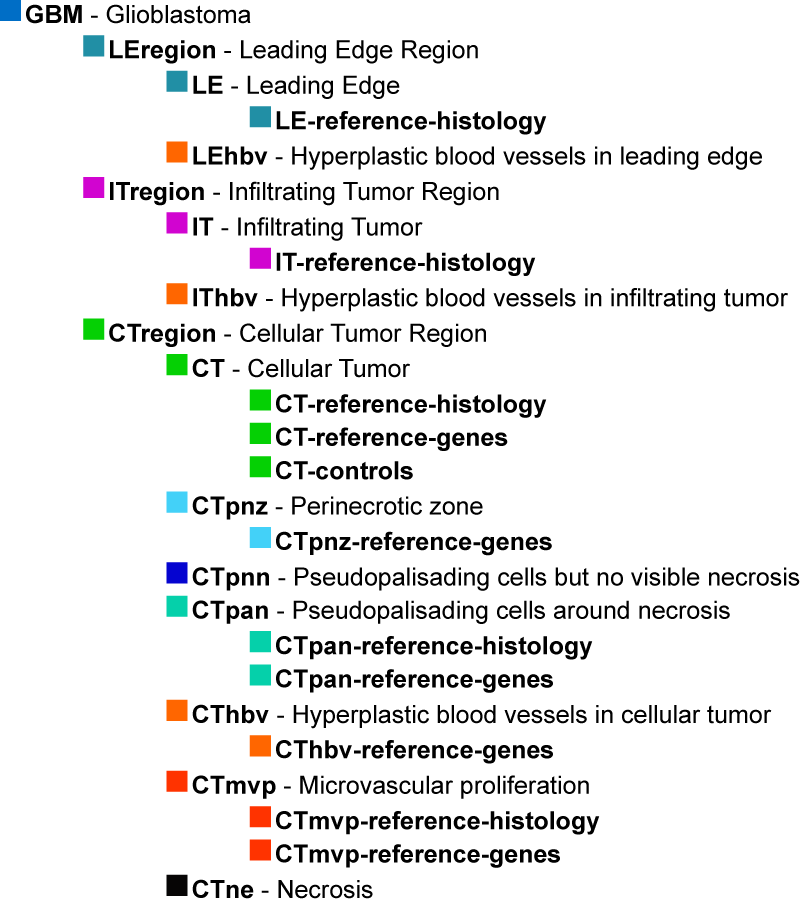
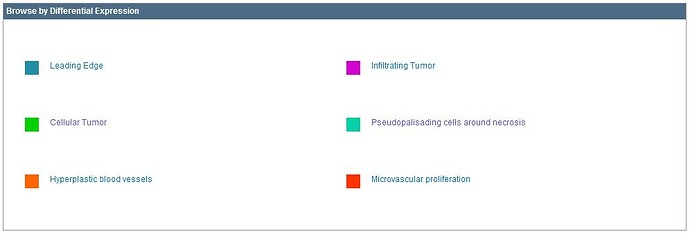
The structural features are commonly identified by neuropathologists in glioblastoma tissue sections stained with Hematoxylin and Eosin (H&E). The major structural regions are Leading Edge (LE), Infiltrating Tumor (IT), and Cellular Tumor (CT).
Within each of these regions, particular structural features can be found such as Microvascular Proliferation (MVP), Pseudopalisading Cells around Necrosis (PAN), Hyperplastic Blood Vessels (HBV), and Necrosis (NE). These features are routinely used to distinguish glioblastoma, or Grade IV glioma, from lower grades of glioma. The feature HBV can be observed occasionally in the LE and IT regions, but it as well as MVP, NE, and PAN are frequently identified in the CT region.
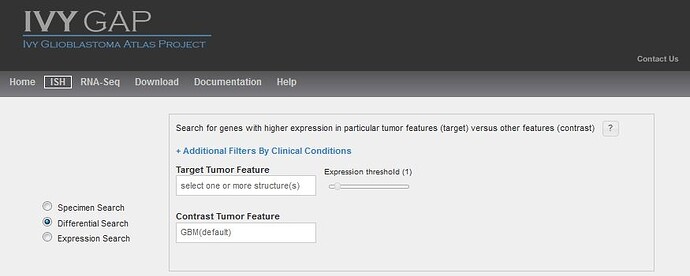
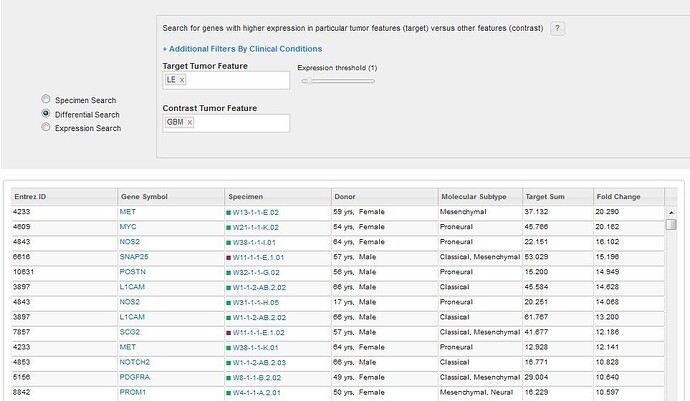
Similar to other Atlases in the Allen Brain Atlas resources, you can search for enriched gene expression in a tumor feature or structure by comparing expression between your target structure and a contrast structure. By default, this search uses all the data as contrast, but changing the contrast structure to more define your search will produce different results. Changing the threshold on a differential search will restrict the list of returned results by excluding any experiment where the expression energy of the target structures is less than threshold.
Several differential searches have already been calculated for you and are accessible by selecting from the “Browse by Differential Expression”.
Performing a differential search will return a list of specimen blocks that include the genes with the highest differential expression levels ranked by fold change in the target structure over the contrast structure.
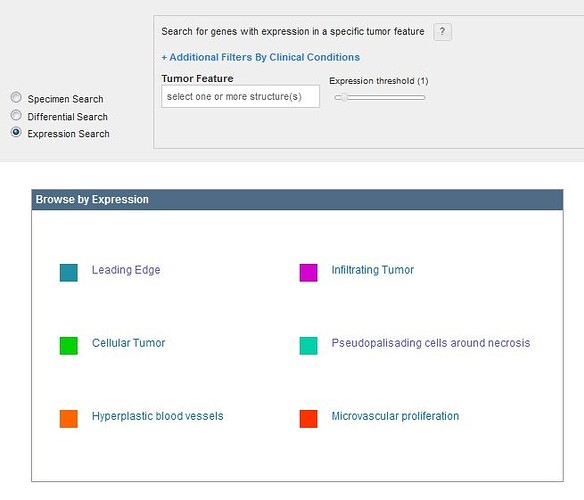
To see gene expression ranked by expression energy within a single glioblastoma structure, use the Expression Search feature. Clicking on a tumor feature from the Glioblastoma Ontology or the “Browse by Expression” box will return a list of genes ranked by the average expression where each column is described below.
When searching for specific images, one of the search tools available to you is to “Filter by Clinical Condition”. Each condition and its significance is outlined below. Listed references can be found in the Overview whitepaper in Documentation.

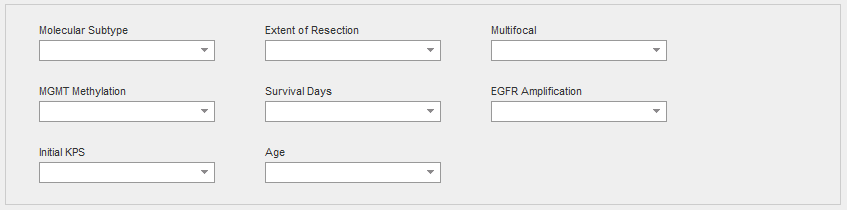
Genomic characteristics of tissue block related to classification of patients. The molecular subtype was determined using all cellular tumor samples from a given tumor. An analysis of each cellular tumor sample was conducted with 840 transcripts from the RNA-Seq data as per Verhaak et al., 2010. For a given tumor, if multiple cellular tumor samples exhibited distinct subtypes, then the tumor was classified as a mixture of its subtypes. The Classical subtype refers to chromosome 7 amplification, specifically EGFR, paired with chromosome 10 loss. The Mesenchymal subtype is noted for focal hemizygous deletions of a region at 17q11.2, containing the gene NF1. The Proneural subtype is defined by alterations of PDGFRA and point mutations in IDh2. The Neural subtype is typified by the expression of neuronal markers such as NEFL, GABRA1, SYT1, and SLC12A5.
Degree to which tumor tissue was surgically removed. Complete resection is associated with increased survival rates (Keles et al., 1999).
Whether a tumor was comprised of single or multiple masses. Increased multifocality is associated with decreased rates of survival (Thomas et al., 2013).
A diagnostic test result referring to whether a tumor’s O-6-methylguanine-DNA methyltransferase DNA repair gene was methylated. Methylated MGMT is associated with increased rates of survival (Hegi et al., 2013).
Patient’s lifespan since initial diagnosis
A diagnostic test result referring to whether a particular tumor’s epidermal growth factor receptor gene had multiple copies. The gene is frequently amplified in glioblastoma (Hobbs et al., 2012).
Karnofsky Performance Status when first diagnosed with first tumor. Karnofsky Performance Status is a measure of functional impairment on a scale of 0-100, 0 referring to dead, 50 to requiring considerable assistance and frequent medical care, and 100 referring to no evidence of disease. High scores are associated with increased survival rates (Ening et al., 2015).
Patient age in years at time of diagnosis. Younger patients tend to have increased survival rates (Ening et al., 2015).
Clicking on a Specimen ID will bring you to the specimen detail page.
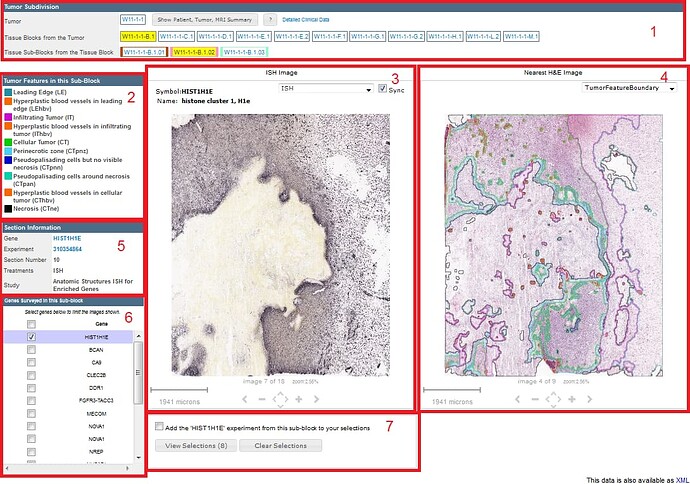
This panel displays information on patient and tumor, and on how the tumor was subdivided into blocks. This panel is arrived at by toggling the “View Patient, Tumor and MRI Summary” button in the Tumor Subdivision section of the Specimen Detail Page.

The tumor names (or IDs) identify tumors across the different resources associated with the Ivy Glioblastoma Atlas Project. In the name W1-1-2, W1 identifies the patient, and -1-2 indicate that this tumor was the second mass removed during the first surgery.
The names of the tumor blocks were assigned as indicated in the Resected Tumor Image, and the names of the sub-blocks reflect their position within the tumor block.
The GBM image viewer allows you to navigate through a series of specimen sub-block ISH images to view at higher resolution. Once a gene image is selected, you can manipulate it with your mouse and use the keyboard or on screen navigation tools to take additional actions.
The gene symbol and gene name are displayed in the title bar along with a dropdown menu and a “Sync” feature to allow you to navigate and keep the adjacent H&E viewer synched with your movements. The dropdown menu allows you to view the ISH image, the Expression Mask, the annotated features, or the annotated feature boundaries. If all the genes are selected in Box #6, the entire specimen sub-block can be viewed using the onscreen navigation tools or the keyboard commands. The current selection is outlined in black.
To view images at high resolution, in the Section Information box, click on the Experiment link; then, click the icon in the upper right corner of the viewer. Use the dropdown menu to select the type of image you want to see.
Alternatively, to view all the images available for a given gene, in the Section Information box, click on the gene symbol link.
Shows the current viewing resolution of the image, in microns. This value dynamically changes as you zoom in/out of the image. You can position the scale bar anywhere on the main image by dragging the scale bar by its ruler.

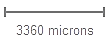
You can toggle the orientation of the scale bar from horizontal to vertical by clicking on the scale bar text.
The Expression mask image display highlights those cells that have the highest probability of gene expression using a heatmap color scale (from low/blue to high/red).

Learn about API For Ivy Glioblastoma Atlas Project with comprehensive guides and examples from Allen Institute for Brain Science.
Learn about Rna Sequencing Data For Ivy Glioblastoma Atlas Project with comprehensive guides and examples from Allen Institute for Brain Science.
Learn about Documentation Ivy Glioblastoma Atlas Project with comprehensive guides and examples from Allen Institute for Brain Science.