Our Science (SEA-AD)
The Seattle Alzheimer’s Disease Brain Cell Atlas (SEA-AD) consortium aims to provide scientific insight on the pathogenesis of Alzheimer’s disease using data generated by SEA-AD and others. Access our latest published work describing cellular and molecular changes in AD progression.
Platform papers
Large studies from the whole SEA-AD consortium describing major data generation efforts and associated knowledge from these data sets.
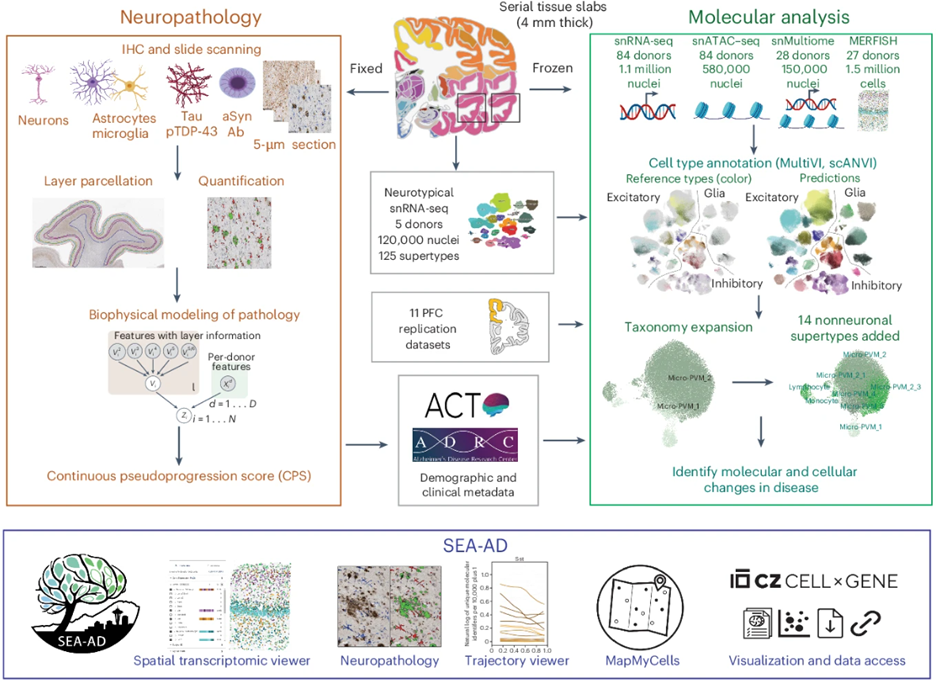
Integrated multimodal cell atlas of Alzheimer’s disease (Gabitto, Travaglini, et al, Nature Neuroscience, 2024)
This paper describes the creation of a massive molecular and cellular atlas of middle temporal gyrus to better frame and understand how Alzheimer’s disease progresses. By analyzing the genetic activity of millions of individual cells, the researchers discovered that the disease actually unfolds in two distinct phases: an early, slow-moving stage where specific Sst+ interneurons are lost and inflammation begins, followed by a later stage of accelerated neuropathology and loss of excitatory neurons and Pvalb+ and Vip+ inhibitory neuron subtypes. These findings were replicated in prefrontal cortex in SEA-AD and other major AD studies.
Focused studies
Other papers with SEA-AD researchers as primary authors focused on SEA-AD data sets or novel computational algorithms.
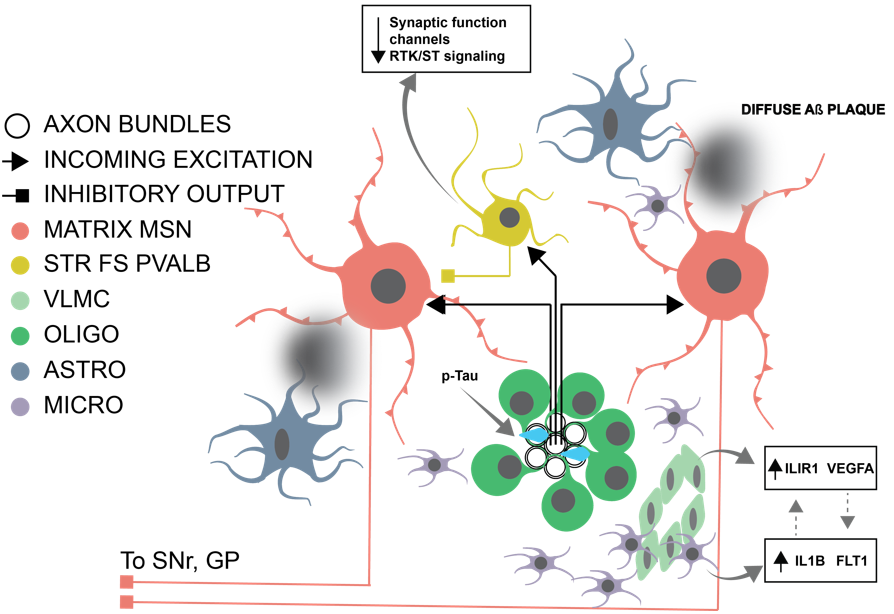
The Caudate Nucleus Exhibits Distinct Pathology and Cell Type-Specific Responses Across Alzheimer’s Disease (Kana et al, bioRxiv, 2026)
This paper represents the characterization of the Alzheimer's pathological spectrum within the caudate nucleus. Utilizing digital pathology, researchers were able to confirm a less pronounced phosphorylated tau and amyloid-β burden when compared to other cortical areas. Phosphorylated tau and amyloid-β were also shown to localize separately into white and gray matter, respectively. Both, in part, explain the lack of neuronal cell death observed in the caudate even in high burden parts of the spectrum. However, researchers were able to identify glimmers of a global phosphorylated tau response in microglia.
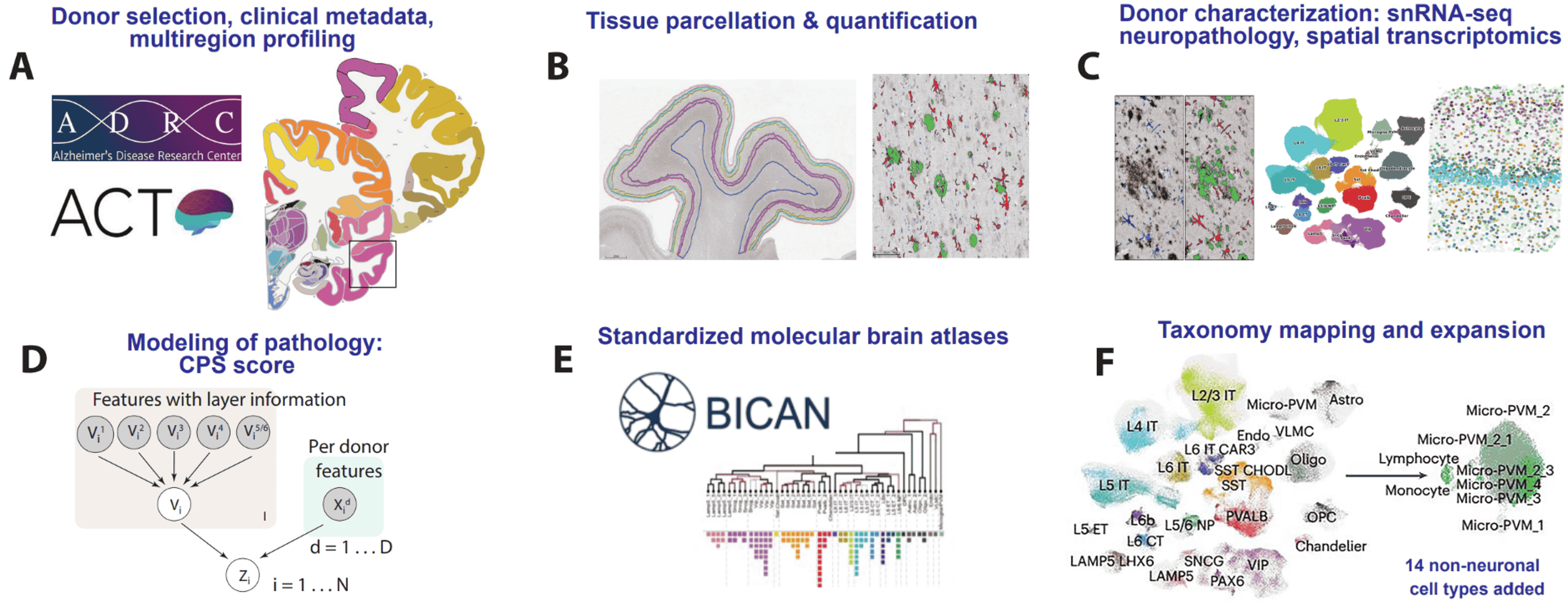
SEA-AD is a multimodal cellular atlas and resource for Alzheimer’s disease (Hawrylycz et al, Nature Aging, 2024)
This commentary on Gabitto, Travaglini, et al 2024 presents SEA-AD as a multifaceted open community resource designed to identify cellular and molecular pathologies that underlie Alzheimer’s disease. Integrating neuropathology, single cell and spatial genomics, and longitudinal clinical metadata, SEA-AD is a unique resource for studying the pathogenesis of Alzheimer’s and related dementias.
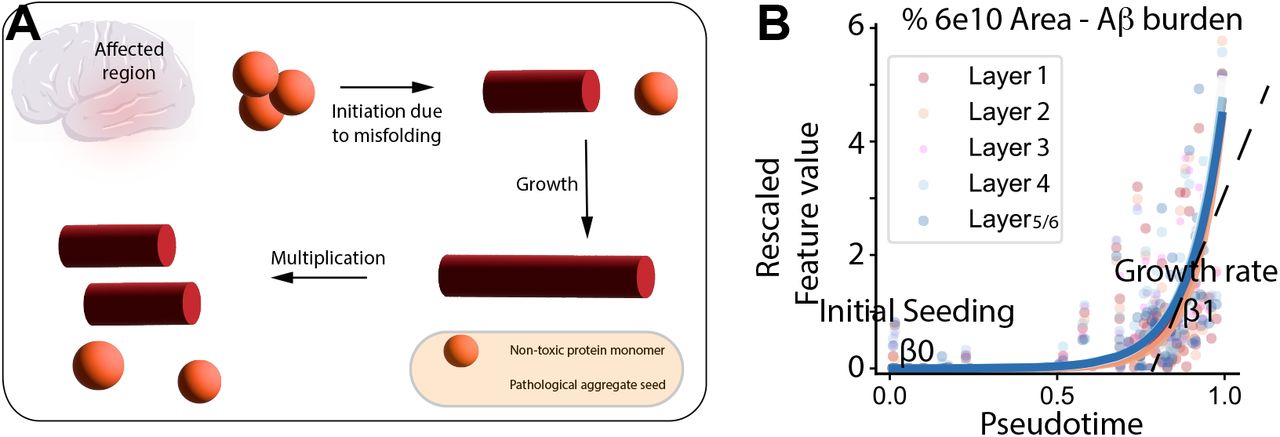
B-BIND: Biophysical Bayesian Inference For Neurodegenerative Dynamics (Agrawal et al, bioRxiv, 2024, accepted at the Annals of Applied Statistics)
This paper introduces B-BIND, a new mathematical framework designed to track how Alzheimer’s disease progresses by analyzing the buildup of various pathological proteins. Because researchers often only have "snapshots" of the brain from different individuals, this model uses a "pseudotime" scale to rank patients along a continuous timeline of disease severity. Ultimately, this framework lays the groundwork for identifying the hidden biological stages of neurodegeneration and the specific cellular changes that drive the disease over time.
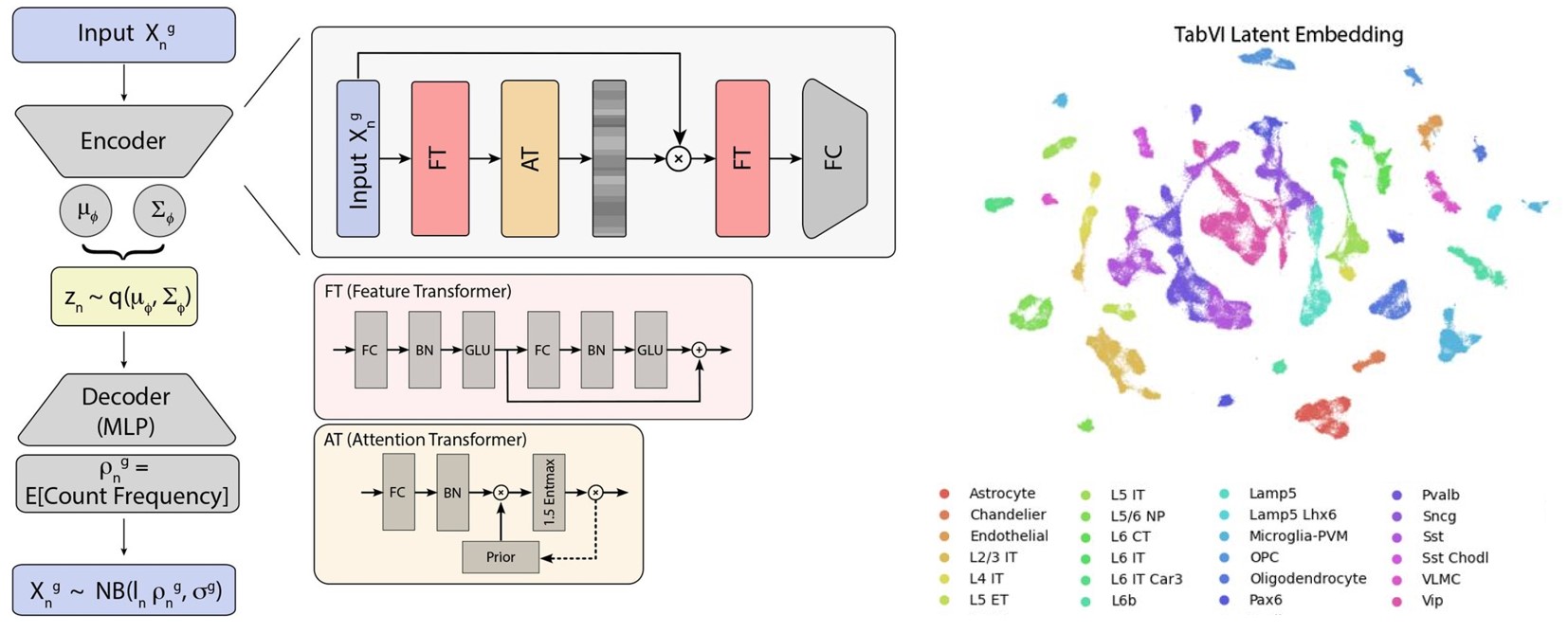
TabVI: Leveraging Lightweight Transformer Architectures to Learn Biologically Meaningful Cellular Representations (Chandrashekar et al, bioRxiv, 2026)
This paper introduces TabVI, a lightweight AI model designed to analyze single-cell genomic data more effectively than standard language-based AI. By adapting "transformer" technology—the same tech behind ChatGPT—to better fit the non-sequential, hierarchical nature of genes, TabVI can more accurately identify cell types and biological patterns. This tool provides a more efficient and interpretable way for researchers to study how diseases like Alzheimer’s affect individual cells across large datasets.
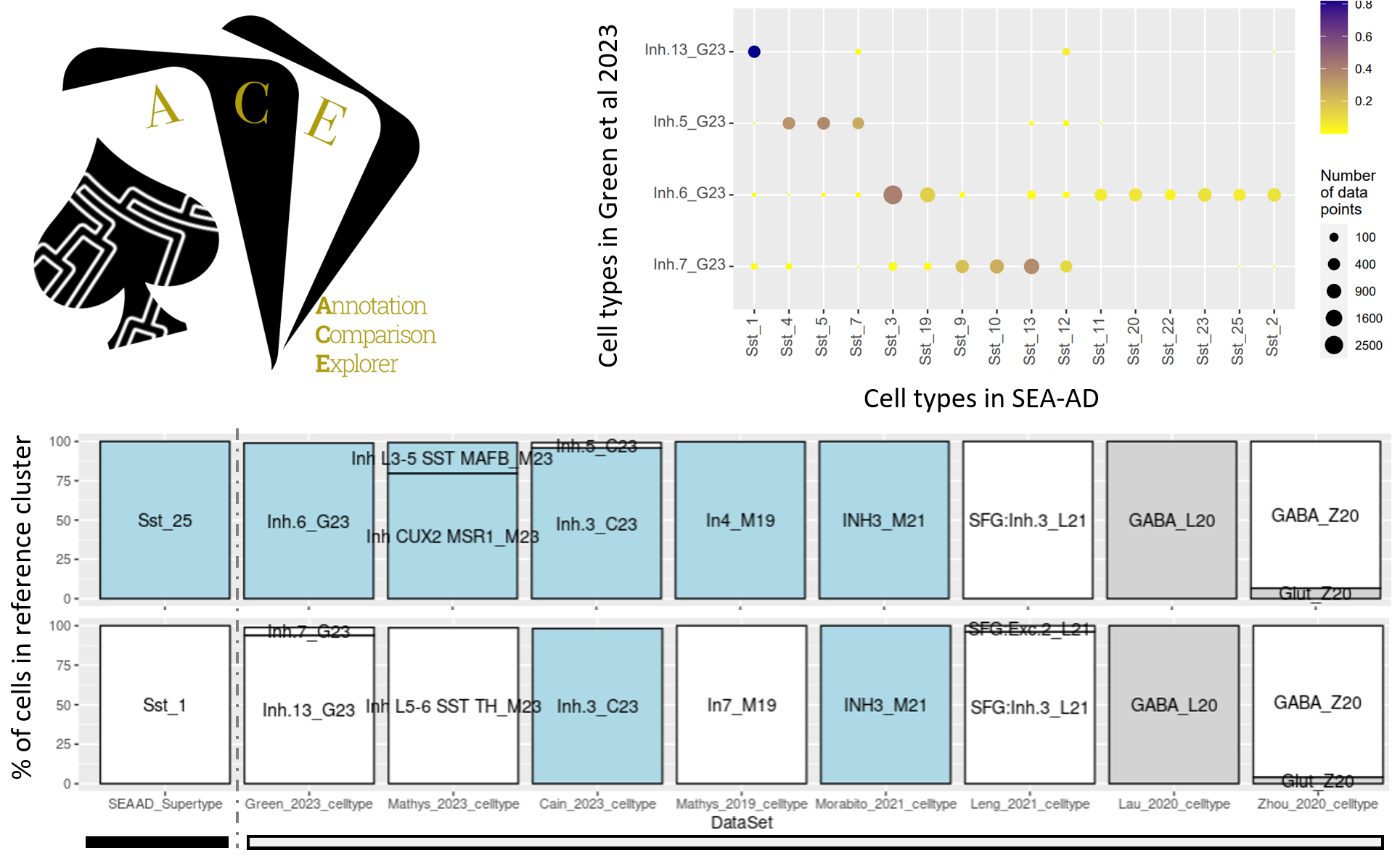
Annotation Comparison Explorer (ACE): connecting brain cell types across studies of health and Alzheimer’s Disease (Miller et al, bioRxiv, 2025)
This paper introduces Annotation Comparison Explorer (ACE), a web-based tool developed by the Allen Institute to address the challenge of comparing cell-type classifications across different brain studies. ACE allows researchers to map their own single-cell data to established taxonomies, such as the Seattle Alzheimer’s Disease Brain Cell Atlas (SEA-AD), and compare annotations like donor demographics and disease-related changes. By applying ACE to multiple published Alzheimer's datasets, the authors identified consistent signatures of the disease, such as a decrease in specific somatostatin interneurons, demonstrating the tool's utility in unifying diverse datasets to advance the understanding of brain health and neurodegeneration.
Collaboratory studies
Projects led by other folks in the AD community, but with significant SEA-AD input or using SEA-AD data resulting in co-authorship by SEA-AD researchers.
Somatic cancer driver mutations are enriched and associated with inflammatory states in Alzheimer’s disease microglia (Huang et al, bioRxiv, 2024, accepted at Cell)
Using transcriptomic datasets (including SEA-AD), this study reveals that somatic mutations—genetic changes occurring after birth—are twice as common in Alzheimer’s brains than in healthy, age-matched controls. These mutations are concentrated in the brain's immune cells (microglia) and often affect "cancer driver" genes, causing the cells to multiply and trigger inflammation. This suggests that Alzheimer's may be driven by a tumor-like process where mutated immune cells expand throughout the brain and accelerate neurodegeneration.
Read the paper | Use RNA-MosiacHunter
Single Cell Landscape of Sex-specific Drivers of Alzheimer’s Disease (Wu et al, Alzheimer’s & Dementia, 2025)
This study analyzed single nucleus transcriptomics data (including from SEA-AD) to understand how Alzheimer's disease affects men and women differently at a genetic level. Researchers found female-specific gene associations providing "protective" effects in neurons of dorsolateral prefrontal cortex, while specific inflammatory signals in microglia may increase risk for protein buildup in women. These findings highlight that the molecular path to cognitive decline is sex-specific, pointing toward new, targeted genes for future treatment research.
Read the paper | Access the SEA-AD data | Access the ROS/Map data
CRISPR screens in iPSC-derived neurons reveal principles of tau proteostasis (Samelson et al, bioRxiv, 2025)
This study utilizes genome-wide CRISPRi screens in human iPSC-derived neurons to identify cellular factors regulating the accumulation of tau aggregates, a primary driver of Alzheimer’s disease. The researchers identified the ubiquitin ligase CUL5-SOCS4, alongside pathways such as UFMylation and mitochondrial function, as critical modifiers of tau stability and potential therapeutic targets. Furthermore, the team leveraged SEA-AD data to validate these findings, demonstrating that these tau-regulating genes exhibit altered expression within the specific cell types most vulnerable to Alzheimer’s in human donors.
Read the paper | Explore CRISPR resources
Studies citing SEA-AD
Recent studies citing SEA-AD publications or data, illustrating SEA-AD's research community impact
Title
Author
Year
Source
Type
Abdel-Haleem et al
2026
Brain
Journal article
Almeida et al
2026
Neural Regeneration Research
Journal article
Chaubey et al
2026
Cell Reports Medicine
Journal article
Chu et al
2026
Free Radical Biology and Medicine
Journal article
Dalley et al
2026
Preprint
Dharshini et al
2026
Nature Communications
Journal article
Duggan et al
2026
Molecular Neurodegeneration
Journal article


Explore our Programs
The Allen Institute Institute for Brain Science is active in a wide variety of areas to accelerate progress towards understanding the brain. Find choice resources below.


