SEA-AD Data and Downloads
Seattle Alzheimer's Disease Brain Cell Atlas (SEA-AD) Study Documentation, Data, and Downloads
The Seattle Alzheimer’s Disease Brain Cell Atlas (SEA-AD) consortium strives to gain a deep molecular and cellular understanding of the early pathogenesis of Alzheimer’s disease. This page includes links to access and download raw and processed data generated as part of SEA-AD, along with documentation describing aspects of data generation from cohort selection to tissue processing, to detailed protocols for collecting and analyzing data from each modality.
Access and Download Data
SEA-AD includes data from multiple brain regions and data modalities. All of the data included in our public resources, along with additional data not yet fully analyzed is freely and publicly available online. This graphic to the right summarizes all data currently available:
Values indicate the number of SEA-AD donors. Note that snOmics data from 3-5 reference donors is also available for most regions. White boxes denote data being collected but not yet available, while gray boxes denote no data collection planned. Abbreviations: MTG, middle temporal gyrus; A9 (also called PFC), prefrontal cortex; STG, superior temporal gyrus; V1C, primary visual cortex; MEC/LEC/EC, medial/lateral entorhinal cortex; HIP, hippocampal formation; ITG, Inferior Temporal Gyrus; AnG, angular gyrus; FI, frontoinsular cortex; CaH, head of the caudate nucleus.
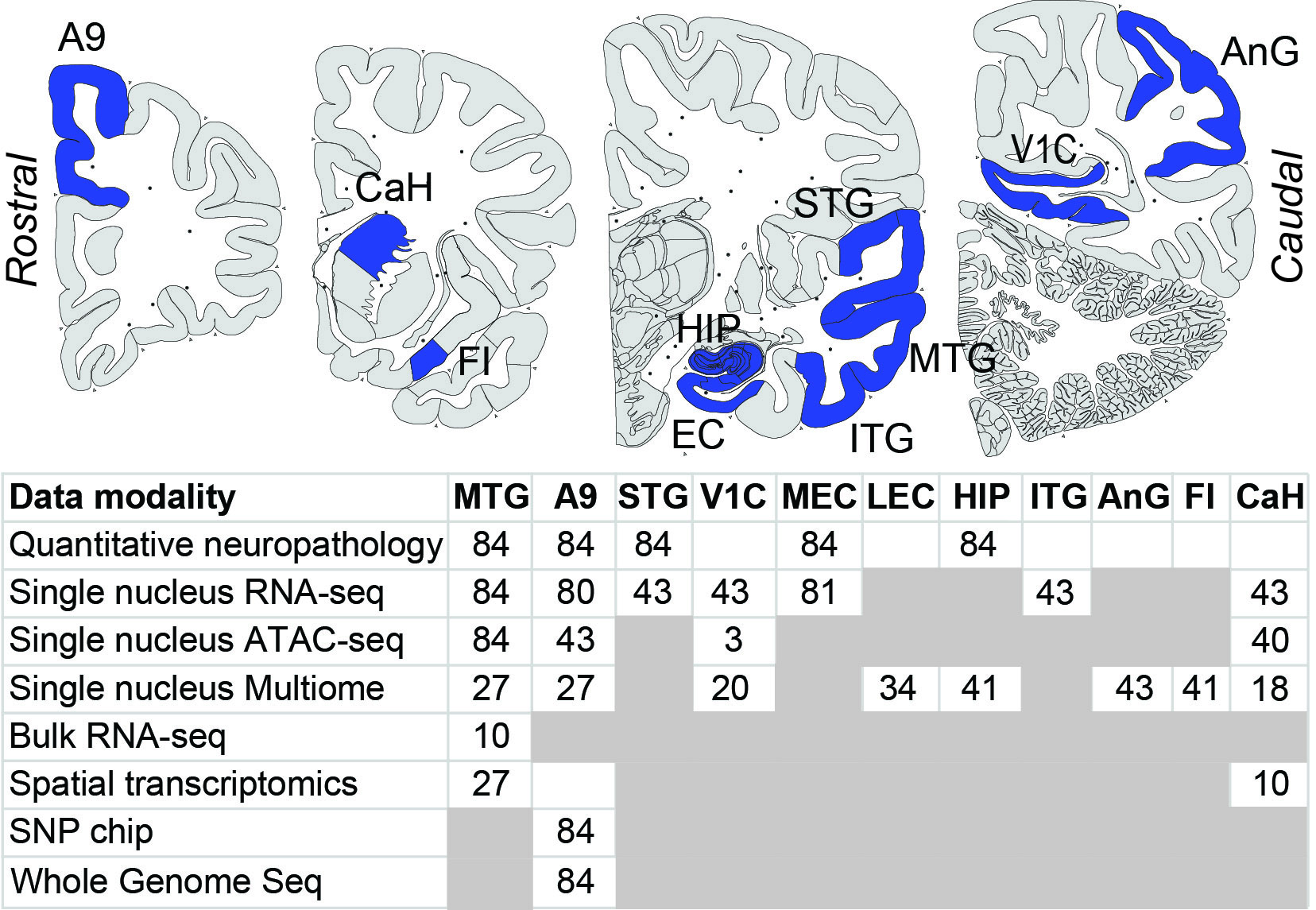
Data
The table below includes a series of files or links out to all SEA-AD data. We provide these data to the research community. Accessing, downloading, and/or using these files must be done in accordance with the Allen Institute Terms of Use and the terms of our external partners. To use our resources, you must cite them in accordance with our Allen Institute Citation Policy. To cite a dataset, cite both the primary publication and the specific dataset. Citation of the primary publication, where one exists, is required when citing the data resource in a scientific publication
File
Size
Type
Description
44KB
.xlsx
Donor metadata (demographic, clinical, cognitive, and neuropathological data)
link
Link to Sage Bionetworks AD Knowledge Portal SEA-AD study page. Controlled access to raw 10x snRNASeq, snATACseq, and Multiome data generated by the SEA-AD study.
link
Link to Sage Bionetworks AD Knowledge Portal SEA-AD Integrative Analysis Center (IAC) study page. Controlled access to single cell genomics datasets and associated representations of disease pathology across the AD/ADRD community through uniform re-processing, data harmonization and mapping to highly annotated BRAIN Initiative Cell Atlas Network (BICAN)/SEA-AD cell classifications.
link
Link to processed single cell data on AWS. Open access to processed 10x snRNAseq and snATAC-seq data.
13KB
.xlsx
Information on the number of available ante-mortem brain scans for donors in the SEA-AD cohort. Access to scans requires contact with the ACT study and/or UW ADRC.
24KB
.xlsx
Luminex data: ABeta and Tau measurements for Middle Temporal Gyrus (MTG).
253KB
.csv
Measurements of Abeta, pTau, pTDP43, a-synuclein, Neun+ cells, IBA1+ cells, and GFAP+ cells from quantitative analysis of stained neuropathology images from Middle Temporal Gyrus (MTG).
5KB
.csv
Measurements of Abeta,pTau, and TDP-43 from quantitative analysis of stained neuropathology images from Caudate Nucleus (Ca) and accompanying Continuous Pseudo-progression Score (CPS) measurements.
link
Link to neuropathology image files on AWS. Download raw and quantified (stained images annotated and masked via the HALO image analysis software from Indica Labs).
link
Link to spatial transcriptomics data: Multiplexed Error-Robust Fluorescence in situ Hybridization (MERFISH) Vizgen MERSCOPE data, and 10x Genomics Xenium data on AWS.
233MB
.csv
Conversion table between cell ids used in the ABC Atlas and cell labels used in all other locations for single cell and MERFISH data sets in MTG and prefrontal cortex (A9).
71KB
.xlsx
Available harmonized cognitive scores for the SEA-AD cohort. The dataset includes composite scores for memory, executive functioning, language, and visuospatial ability at each visit and their slope of overall. Each of the domains was psychometrically co-calibrated across the Adult Changes in Thought (ACT) study, the Alzheimer’s Disease Neuroimaging Initiative (ADNI), the Religious Orders Study (ROS), the Memory and Aging Project (MAP), and the University Of Pittsburgh (PITT) study.
link
Link to the landing page for the Mouse Mesoscale Connectivity in Aging and Alzheimer’s Disease project (unaffiliated with SEA-AD). This project involved quantifying brain-wide axonal projections and amyloid pathology in mouse models of AD and their aged littermates.
3GB
link
Link to AWS bucket for the microglia nuclei data. The data are released as an AnnData file and contains gene expression information on 240,651 nuclei (the vast majority of which are Microglia) for 36,601 features/genes contained with the 10x Genomics 2020-A human reference. Alignments for singleome RNAseq (10x v3.1 3') and multiome (10x) were performed identically with cellranger (v6.1.2) and cellranger-arc (v2.0.0), respectively.
Access the white papers
White papers’ documenting various aspects of SEA-AD cohort and data generation. Links and associated descriptions are shown in the table below.
File
Size
Type
Description
514KB
Graphs summarizing the SEA-AD cohort's age at death, sex, APOE4 status, cognitive status, CERAD score, ADNC score, Braak stage, and Thal phase.
95KB
Methods for 10x snRNASeq, snATACseq, and Multiome data generation and RNA Integrity Number (RIN) determination
153KB
Methods for transcriptomics data analysis. Information on development of web tools, which present transcriptomic data.
123KB
Information on clinical AD detection and consensus clinical diagnosis generation.
136KB
Information on clinical AD detection and consensus clinical diagnosis generation.
108KB
Citation for dissection, ex vivo MRI, tissue preparation, and neuropathological analysis.
217KB
Methods information and tutorial for processed single cell profiling data available on AWS
764KB
Methods information and tutorial for neuropathology images and image analysis data available on AWS
764KB
Translation table between initial cell type names used in Comparative Viewer and CELLxGENE and official names used everywhere else.



Explore our Programs
The Allen Institute for Brain Science is active in a wide variety of areas to accelerate progress towards understanding the brain. Find choice resources below.

